-
Barnyard millet (Echinochloa esculenta) is garnering attention for its potential to contribute significantly for food and nutritional security, particularly in Asia[1]. It ranks as the fourth most produced minor millet and India leads globally in barnyard millet production, covering the largest area (0.146 Mha) and yielding the highest output (0.147 Mha) with an average productivity of 1,034 kg·ha−1 over the past three years[2]. E. esculenta, known as Japanese barnyard millet, is valued for its fast growth, adaptability to various soil and water conditions, and nutritional grain profile, making it an excellent choice for forage and food[3]. Moreover, E. frumentacea, or Indian barnyard millet, shares similar traits but is often preferred in traditional farming systems in South Asia due to its hardiness and ease of cultivation[4]. In crop improvement, E. esculenta's rapid growth and resilience are leveraged to enhance stress tolerance and yield in other cereal crops[5,6]. Barnyard millet emerges as a beacon of hope for regions grappling with unpredictable weather patterns, and its secret lies in its remarkably short life cycle, maturing in just 60−80 days[7]. This swiftness allows farmers to double-crop, squeezing in two harvests within a single season[8].
Even more crucially, the short cycle enables them to avoid droughts by planting just before the rains and harvesting before potential dry spells[9]. Additionally, the nutritional benefits of barnyard millet, including being rich in protein, carbohydrate and dietary fiber, low glycemic index, gluten-free, abundance of micronutrients like iron and zinc, good source of vitamin B, and rich in antioxidants, position it as a valuable crop for addressing nutritional deficiencies, particularly in hilly and tribal communities[10].
Moreover, Fig. 1 categorizes different biological data types and resources, along with the number of records available for each in NCBI (National Center for Biotechnology Information) database as of 20th July 2024 for Japanese barnyard millet (E. esculenta) (Taxonomy ID: 121770)[11]. For literature, sources include Bookshelf, PubMed, and PubMed Central, totaling 168 records[11]. Genes are covered by the Gene and PopSet databases with 142 records, while Proteins are split into Identical Protein Groups and Protein databases with 91 and 203 records respectively[11]. Genomes are sourced from BioProject and Taxonomy with five records, BioSample with 165 records, Nucleotide with 69 records and the Sequence Read Archive with 164 records. Lastly, the PubChem database includes 21 BioAssay records[11].
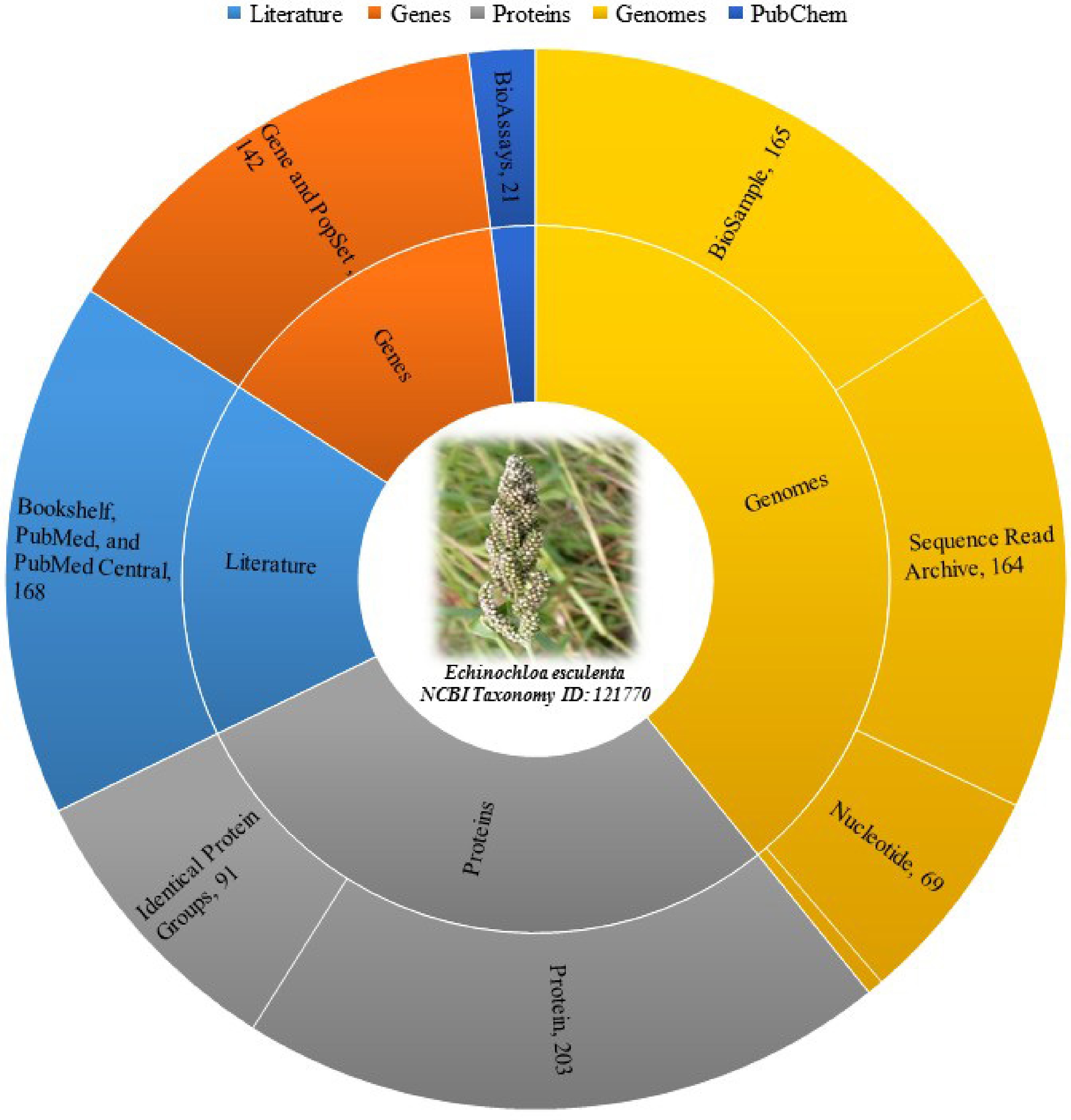
Figure 1.
Distribution and abundance of different types of biological data of Japanese barnyard millet (E. esculenta) across various databases as of 20th July 2024 [11].
Moreover, the genetic improvement of barnyard millet holds significant potential but requires concerted research efforts to overcome existing challenges[12]. However, genome research in barnyard millet is still in its early stages and lags significantly behind other minor millets, and this is primarily due to the complex nature of its genome (2n = 6x = 54, hexaploid)[6]. Enhanced genetic characterization, effective utilization of germplasm resources, and advanced breeding techniques, including interspecific hybridization, are essential for unlocking the full potential of this underutilized crop[5]. The limited genetic diversity observed within cultivated varieties, compared to their wild relatives, further complicates breeding efforts aimed at enhancing desirable traits like yield, disease resistance, and abiotic stress tolerance[13].
On the positive side, recent advancements in genomics have opened new avenues for the genetic enhancement of barnyard millet[5]. The release of genome and transcriptome sequences for both wild and cultivated Echinochloa sp. has facilitated a better understanding of the genetic basis of important agronomic traits[9]. The identification of quantitative trait loci (QTL) and specific genes associated with these traits is crucial for the development of improved varieties[9]. Furthermore, interspecific hybridization between E. esculenta and E. frumentacea has shown promise, despite challenges related to seed sterility in hybrid plants[14]. This hybridization can potentially introduce desirable traits from one species to the other, thereby broadening the genetic base and enhancing the crop's overall performance[15].
Despite nutritional advantages, barnyard millet remains underutilized[9]. The genetic improvement of barnyard millet faces several challenges and presents numerous prospects. One of the main challenges is the limited genetic and genomic resources available for this crop[7]. Although recent efforts have led to the development of core collections and the identification of single-nucleotide polymorphisms (SNPs) within these collections, there is still a significant gap in understanding the full genetic architecture of barnyard millet[16]. Moreover, integrating advanced molecular and biotechnological tools with conventional breeding approaches can significantly enhance the agronomic traits of E. esculenta[17].
Techniques such as genome editing with CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) - Cas9 (CRISPR-associated protein 9) can precisely target and modify genes associated with important agronomic traits in barnyard millet[18]. Marker-assisted selection (MAS) allows for the efficient identification and incorporation of desirable traits by using molecular markers linked to key agronomic characteristics in barnyard millet[19]. Combining these molecular tools with traditional breeding ensures the development of superior E. esculenta varieties with enhanced performance and resilience[20−22].
Therefore, the main focus of this review is to explore how these advanced tools can be effectively integrated with conventional methods to improve the agronomic traits of E. esculenta, emphasising the potential for developing high-yield, stress-resistant barnyard millet varieties to enhance food security and agricultural sustainability.
-
Barnyard millet, despite its name, is a grain belonging to the grass family Poaceae[22]. While its exact origin remains unclear, it's believed to have been domesticated in India or Southeast Asia. Classified within the genus Echinochloa, it shares some relatives with other millets but possesses a unique genetic makeup[23]. Moreover, barnyard millet is a significant crop primarily cultivated in semi-arid tropical regions of Asia and Africa appreciated for its drought tolerance, rapid maturation, and nutritional benefits[24]. The genus Echinochloa comprises multiple species, with the two major cultivated types being E. frumentacea (Indian barnyard millet) and E. esculenta (Japanese barnyard millet)[25]. The origins, classification, and genomic relationships of barnyard millet can be understood through a synthesis of genetic and genomic research[24].
Barnyard millet has a complex origin involving multiple species and subspecies[13,22]. The genomic relationship of barnyard millet species has been explored using various molecular markers[16]. A study utilizing whole-genome genotyping-by-sequencing on a core collection of 89 barnyard millet accessions identified several thousand SNPs segregating within the collection[16]. This study revealed four population clusters within E. colona and three within E. crus-galli, suggesting that the genetic diversity within these species is substantial and likely influenced by geographic factors[16]. Additionally, the use of RAPD (Random Amplified Polymorphic DNA) primers confirmed the genetic diversity among 21 accessions of the Echinochloa spp. complex, classifying them into two morphological races, E. frumentacea and E. colona, with a high degree of molecular diversity[26].
Moreover, interspecific hybridization between E. esculenta and E. frumentacea offers potential for genetic enhancement[26]. A successful hybrid between E. esculenta cultivar PRJ 1 and E. frumentacea cultivar ER 72 is also reported, confirmed through rice SSR markers[15]. Although the hybrid plants exhibited vigor and disease resistance, they were sterile, highlighting challenges in utilizing interspecific hybrids for breeding programs[15]. Therefore, the genetic characterization of barnyard millet is crucial for breeding and improvement programs. For instance, association analysis in germplasm and F2 populations have shown positive direct effects of traits like stem girth, ear head weight, and plant height on grain yield[27]. Moreover, understanding its genomic relationships with other Echinochloa species is crucial for breeding programs to develop improved barnyard millet varieties with enhanced yields and desirable traits[23].
-
The phylogeny of barnyard millet has been the subject of multiple studies focusing on genetic diversity, population structure, and evolutionary relationships among different species and accessions[16]. However, these studies identify thousands of SNPs and revealed distinct population clusters, and interestingly, these genetic clusters correlated more with geographic origin rather than morphological classification, highlighting the influence of geographic isolation on genetic diversity[28]. Another significant contribution to understand the phylogeny of Echinochloa species comes from the analysis of chloroplast DNA (cpDNA) sequences from non-coding regions (trnT-L-F)[29], and the findings suggest that domestication and adaptation to different environments occurred after the species diverged in Asia[28].
Further phylogenetic insights are provided by the complete chloroplast genome sequencing of Indian barnyard millet (E. frumentacea), and this study revealed that E. frumentacea diverged from its close relatives E. oryzicola and E. crus-galli approximately 1.9–2.7 million years ago[30]. However, morpho-agronomic trait analysis has contributed to our understanding of barnyard millet phylogeny[31]. Lastly, the assessment of genetic diversity using molecular markers further elucidates the phylogenetic relationships within and between E. crus-galli and E. frumentacea[32]. However, the phylogeny of barnyard millet is complex and influenced by geographic, genetic, and morphological factors[33]. Moreover, the phylogeny of barnyard millet is characterized by significant genetic diversity and complex evolutionary relationships, and studies utilizing whole-genome sequencing, interspecific hybridization, and molecular markers have provided a deeper understanding of these relationships, offering valuable insights for crop improvement[34].
-
Germplasm resources in barnyard millet hold significant potential for crop improvement, particularly in terms of enhancing traits such as drought tolerance, nutritional quality, salinity tolerance, and disease resistance[35]. Several studies have characterized the genetic variability and diversity within barnyard millet germplasm, providing valuable insights for breeders and researchers[35]. This genetic diversity is crucial for the development of improved cultivars with desirable traits[26]. The formation of a core set in barnyard millet germplasm using data on 24 morpho-agronomic traits has been instrumental in managing and utilizing the vast genetic resources available[16]. This core set captures most of the available diversity and serves as a representative sample for detailed evaluation and breeding programs[31].
Similarly, another study characterized 494 barnyard millet germplasm for quantitative traits, revealing significant variation for most traits and highlighting traits like days to 50% flowering, plant height, and days to maturity as key contributors to total variance[36]. Moreover, barnyard millet germplasm has been screened for resistance to grain smut with accessions showing varying levels of susceptibility, and the study suggests that smut resistance and grain yield can be improved simultaneously through proper breeding strategies[37]. Additionally, the crop's tolerance to salinity has been investigated, with certain genotypes demonstrating the ability to withstand up to 200 mM salt concentration during germination, and it is found that, this tolerance is associated with traits such as higher relative water content and enzyme activity under salt stress[38]. The genetic variability and association studies under sodic soil conditions have identified traits like days to 50% flowering, days to maturity, ear width, and thousand-grain weight as important selection indices for enhancing grain yield[39].
The characterization of genetic diversity and the formation of core collections facilitate the efficient utilization of barnyard millet germplasm in breeding programs aimed at developing high-yielding, stress-tolerant, and disease-resistant cultivars[40]. While the exact number of barnyard millet germplasm accessions globally remains around 8,000 conserved in various gene banks, such as Vivekananda Parvatiya Krishi Anusandhan Sansthan (VPKAS), India, Indian Institute of Millets Research (IIMR), India, National Institute of Agrobiological Sciences (NIAS), Japan, and Consultative Group on International Agricultural Research (CGIAR), including the International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), India, recent efforts focus on optimizing their use and maintenance[9]. The extensive genetic resources available in barnyard millet germplasm provide a solid foundation for crop improvement[4,41].
-
Genome sequencing in barnyard millet has made significant strides in recent years, providing valuable insights into the genetic makeup and potential for crop improvement in this under-researched species[15,16]. A groundbreaking study on the genetic characterization of a global barnyard millet germplasm collection utilized whole-genome genotyping-by-sequencing to identify several thousand SNPs in 89 accessions, providing a robust framework for breeders to enhance barnyard millet for smallholder farmers[16]. However, researchers in China successfully released and annotated the whole genome sequence of weedy E. crus-galli, and the genome, sequenced at a depth of 171x, was estimated to be 1.27 Gb, covering approximately 90.7% of the predicted size[9]. Moreover, the study identified 4,534 contigs ranging from 1 kb to 11.7 Mb, with gene annotations revealing 108,771 protein-coding genes, 785 miRNAs, 514 Mb of repetitive elements, and various non-coding RNAs[9]. NCBI resources for Echinochloa species included 1,246 nucleotide sequences, 822 gene sequences, 2,468 protein sequences, and other genomic data, with E. crus-galli leading in sequence availability[9]. Despite this, cultivated barnyard millet species E. frumentacea and E. esculenta had significantly fewer sequences, representing only 4% of the total[9].
Further insights were gained from the complete chloroplast genome sequence of Indian barnyard millet, E. frumentacea[30]. The chloroplast genome, which is 139,593 bp in length with a typical quadripartite structure, includes 112 individual genes, and these genes consist of 77 protein-coding genes, 30 tRNA genes, four rRNA genes, and one conserved open reading frame, with an overall GC content of 38.6%[30]. Phylogenetic analysis indicated that E. frumentacea diverged from its close relatives, E. oryzicola and E. crus-galli, approximately 1.9–2.7 million years ago, highlighting the evolutionary history and genetic uniqueness of this species[30]. Moreover, genomics and genetic studies in barnyard millet are rapidly advancing, providing a wealth of data and opportunities for crop improvement[14]. However, in a recent study, the transcriptome profiling of barnyard millet was performed during grain development to reveal the genomic insights into iron accumulation, which has an insight towards genome sequencing[5,8].
-
Molecular markers, specifically nucleotide sequences, play a crucial role in genetic diversity studies, linkage map construction, and marker-assisted selection in crop plants[9]. Early research utilized RAPD markers to study genetic diversity and phylogeny among Echinochloa species, effectively distinguishing between cultivated and wild progenitors[42,43]. Although RAPD markers provided insights into genetic diversity, their low polymorphism levels were noted in various studies[44]. Moreover, Amplified Fragment Length Polymorphism (AFLP) markers later showed greater efficacy in revealing genetic diversity due to their higher allele production per primer[5]. Advancements in sequencing technologies introduced sequence-based markers like SRAP (Sequence-Related Amplified Polymorphism); SSRs (Simple Sequence Repeats); EST-SSRs: (Expressed Sequence Tag-Simple Sequence Repeats); and SNPs, which are more desirable due to co-dominant, reproducible, and highly polymorphic nature[18]. Studies utilizing SSR markers in Echinochloa species have shown the formation of distinct clusters, highlighting the genetic diversity[45−48].
EST markers have proven effective in analyzing genetic diversity, with in silico mining and validation of EST-SSR primers identifying frequent repeat motifs in barnyard millet[49−51]. Additionally, the RAD (Restriction-site associated) approach combined with Illumina DNA sequencing in E. phyllopogon facilitated the rapid discovery of SSR and SNP markers, demonstrating the usefulness of these markers in studying genetic diversity and aiding molecular breeding[52]. Further developments in whole-genome genotyping-by-sequencing (GBS) have identified a significant number of SNPs across various biotypes of Echinochloa species[53]. Moreover, population structure analysis using these SNPs has provided clear genetic differentiation among species, forming distinct clusters[35]. This detailed SNP data enhances our understanding of genetic diversity, origins, and distribution, particularly in herbicide-resistant populations, and supports molecular breeding efforts[54]. Recent studies have continued to use EST-SSR markers to analyze genetic diversity in Indian barnyard millet germplasm, reinforcing the importance of advanced molecular markers in crop genetic research and breeding programs[55]. However, Table 1 summarizes the availability of molecular resources in barnyard millet.
Table 1. Availability of molecular markers in barnyard millet.
Experiment Type of marker Results Ref. Proximate and molecular study 20 Echinochloa frumentacea L. 10 RAPD primers generated 50 bands All 50 bands were polymorphic (100%) having 45 shared and five unique bands; fragment size ranged 212 bp [42] Proximate and molecular study 20 Echinochloa frumentacea L. 10 ISSR primers generated 42 bands 40 bands were polymorphic (90%) with 39 shared one unique bands; fragment size ranged 250 bp [42] Genetic diversity analysis of millet crop Four RAPD primers OPC06, OPC18, OPD13, and OPW04 Two primers OPC06 and OPD13 showed the highest polymorphism (83%) [44] Assessment of genetic diversity in Echinochloa
crus-galli (L.)Eight specific SSR markers 48 alleles were identified; genetic variation among populations (37.01%) [45] Assessment of molecular diversity and proximate composition of barnyard millet, pearl millet and sorghum (five varieties of each) 10 SRAP primer 65 polymorphic bands having 56 shared and nine unique bands with an average of 6.5 bands per primer and 98.75% polymorphism per primer [47] Comparative analysis of whole chloroplast genomes of Echinochloa crus-galli var. crus-galli, E. crus-galli var. zelayensis, and E. glabrescens 139 SSRs Phylogenetic tree between 10 barnyard grass species and other common Gramineae plants, showing new genetic relationships of the genus Echinochloa [46] Validation of ssr markers for barnyard millet obtained by partial genome assembly 46,157 SSRs identified from 11,39,481 contigs 15 SSR markers validated among the 30 barnyard millet accessions [48] Genetic diversity in the barnyard millet 51 EST-SSR markers used 14 primers found polymorphic with 29 alleles, and average PIC of 0.43 [49] Transcriptomic profiling of Echinochloa frumentacea and barnyard grass 30 EST-SSR primer pairs 10 EST-SSR primers were found polymorphic; revealing putative genes involved in drought adaptation and micronutrient accumulation by [50] Inheritance and identification of EST-SSR marker(s) associated with the anthocyanin pigments in barnyard millet 51 EST-SSR markers used Marker BMESSR 39 found associated with the anthocyanin pigment [51] Barnyard millet: EST, SNP markers identified 41 EST sequences 22 microsatellite markers and SNPs identified; one potential SNP and one reliable SNP and two haplotypes obtained [35] Molecular diversity analysis of 48 Echinochloa frumentacea genotypes 182 RAPD marker loci 170 RAPD marker loci (93.40%) found to be polymorphic [55] -
The genetic improvement of barnyard millet is essential for enhancing its productivity, resilience, and nutritional value, making it a key player in sustainable agriculture[4]. This process involves utilizing advanced breeding techniques such as genomic selection, and biotechnological tools to develop superior cultivars with improved traits including higher yield, enhanced resistance to biotic and abiotic stresses, and better nutritional profiles[56]. Despite its potential, the genetic improvement of barnyard millet faces challenges like limited genetic diversity, insufficient genomic resources, and inadequate funding for research[5,8]. Addressing these issues through comprehensive research initiatives and international collaboration can pave the way for the development of robust barnyard millet varieties[15]. However, Fig. 2 illustrates the process of improving barnyard millet cultivars by integrating genetic and omics resources. Genetic resources such as core germplasm, recombinant inbred lines and mutants are utilized alongside genome sequencing to facilitate the identification of molecular markers and genetic improvement techniques like genomic selection and genome editing. Omics resources, including genomics, transcriptomics, and proteomics play a crucial role in phenotyping, genotyping, and ultimately developing enhanced barnyard millet varieties.
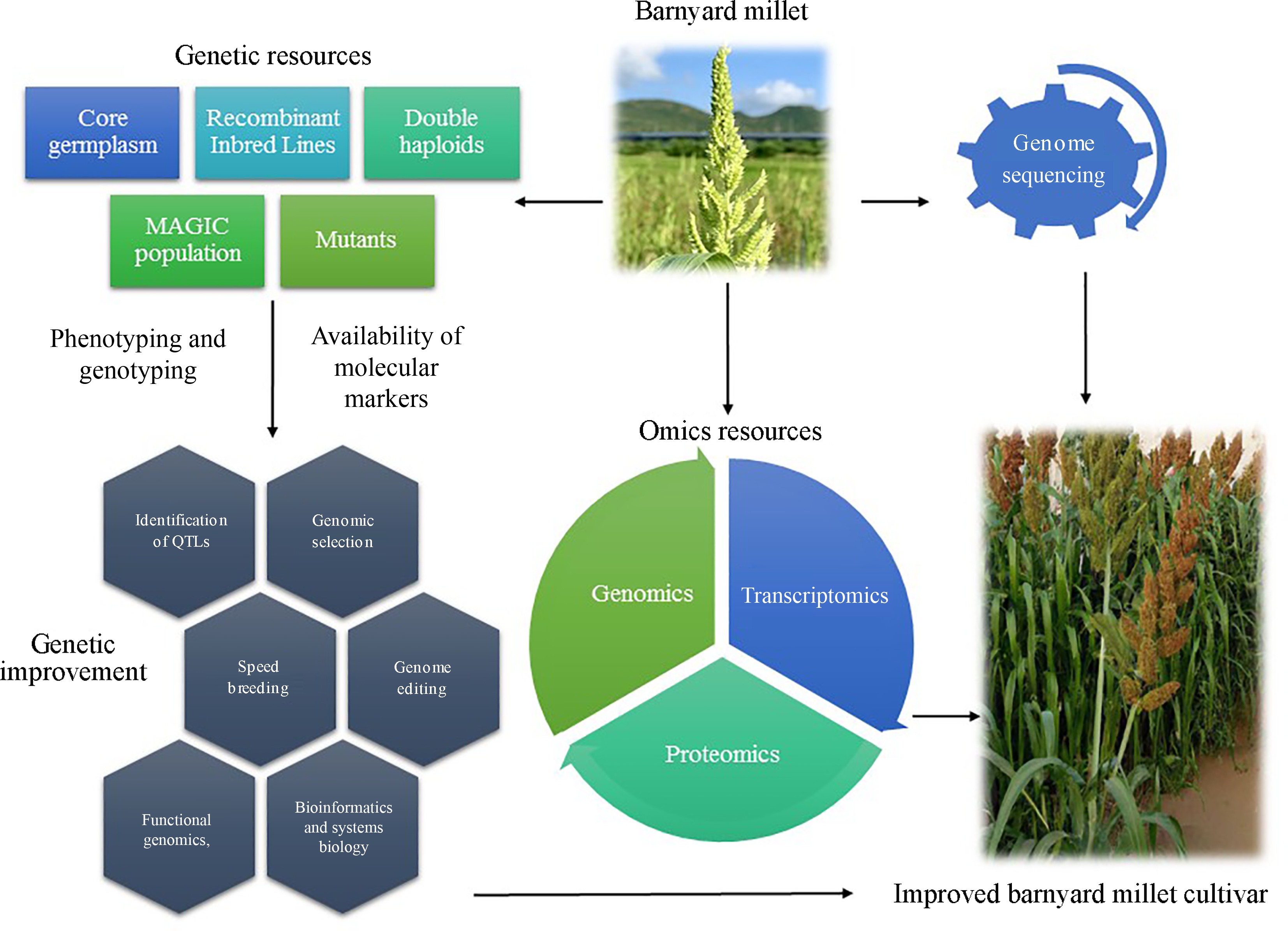
Figure 2.
Process of improving barnyard millet cultivars by integrating genetic and omics resources.
Identification of QTLs and Marker Assisted Selection (MAS)
-
The identification of QTLs and the application of MAS in barnyard millet have been facilitated by the development of molecular markers such as SSRs[45,46] and SNPs[52]. Although many SSR and SNP markers have been created to aid in linkage map construction and QTL mapping, progress has been slower compared to other millets like foxtail and finger millet[55]. Notable studies that identified SNP markers for waxy traits controlled by three loci (EeWx1, EeWx2, EeWx3)[57], and linked the SSR marker BMESSR 39 with anthocyanin pigments using bulk segregant analysis using F2 individuals of ACM 331 × MA 10[58], provide opportunities to facilitate the construction of linkage maps and QTL mapping in barnyard millet followed by MAS[5]. Despite these advancements, barnyard millet genome mapping remains in its initial stages, requiring further investigation to apply these findings to MAS for improved breeding[8].
Genomic selection and speed breeding
-
Barnyard millet's breeding is poised for a breakthrough with the combined power of genomic selection and speed breeding[45]. Genomic selection, unlike traditional methods, analyzes a plant's entire genetic code to predict valuable traits like yield and disease resistance[46]. This allows breeders to identify promising candidates[55]. Speed breeding, on the other hand accelerates the breeding cycle by manipulating the growing conditions to produce multiple generations per year[5,8]. By identifying superior genetics early and rapidly testing them through multiple generations, this combined approach can significantly speed up the development of improved barnyard millet varieties with enhanced yields, disease resistance, and adaptation to local environments[59,60].
Genome editing
-
Barnyard millet, despite its potential as a nutritious and climate-smart crop faces limitations in breeding due to the lack of a complete genome sequence[61]. While traditional breeding methods are ongoing, genome editing offers a promising but nascent approach[62]. CRISPR-Cas, a revolutionary gene editing technique could be a game-changer[63]. However, its application in barnyard millet is still in its early stages[61]. Moreover, chloroplast genomic data can offer significant advantages when implementing the CRISPR/Cas9-based strategy for genetic improvement in barnyard millet. Firstly, chloroplasts possess unique genetic characteristics that can be exploited for genetic engineering[64]. Unlike nuclear DNA, chloroplast DNA is maternally inherited in most plants, which can help contain the spread of transgenes via pollen[63]. This characteristic is advantageous for minimizing gene flow to non-target species and ensuring biosafety in genetically modified crops[65]. Secondly, the efficiency of chloroplast transformation has traditionally been lower than that of nuclear transformation due to the low rate of homologous recombination in chloroplasts[63].
However, recent advancements in CRISPR/Cas9 technology have shown that this can be improved[65]. For instance, using CRISPR/Cas9 to introduce double-strand breaks at specific sites within the chloroplast genome can activate the DNA damage repair mechanism, thereby enhancing the efficiency of homologous recombination. Studies have demonstrated that incorporating CRISPR/Cas9 into the chloroplast transformation system can increase transformation efficiency 6−10 times, which is a substantial improvement[65]. Moreover, CRISPR/Cas9 technology allows for precise genetic modifications, including insertions, deletions, and substitutions, which are crucial for targeted genetic improvement[46].
In the context of barnyard millet, precise editing of the chloroplast genome could be used to introduce traits that enhance photosynthetic efficiency, nutritional value, or stress tolerance, directly impacting crop yield and resilience[64,66]. Additionally, the use of chloroplast genomic data can facilitate multiplex genome engineering, where multiple genes can be targeted simultaneously[63]. This is particularly beneficial for complex traits that are controlled by multiple genes[46]. For example, in cotton, CRISPR/Cas9 has been used to generate mutations in multiple genes concurrently, demonstrating the potential for similar approaches in barnyard millet[46]. Furthermore, chloroplast-targeted CRISPR/Cas9 applications can improve the nutritional quality of barnyard millet by modifying pathways involved in the synthesis of essential nutrients[65]. Given that chloroplasts are the site of important biosynthetic pathways, such as those for amino acids and fatty acids, editing the chloroplast genome provides a direct route to enhancing the nutritional profile of the crop[66]. By successfully utilizing CRISPR-Cas, scientists could directly target genes responsible for desirable traits like higher yield, improved drought tolerance, or resistance to specific diseases, and this would significantly accelerate the development of enhanced barnyard millet varieties, but further research is needed to unlock the full potential of genome editing for this promising crop[61].
Functional genomics, bioinformatics, and systems biology approach
-
Functional genomics is a powerful approach to understanding the roles of genes in complex biological systems and can be highly beneficial for improving crops such as barnyard millet[67,68]. Applying RNAi techniques to barnyard millet could similarly facilitate the identification of key genes involved in stress responses, growth, and yield[69,70]. Another critical tool in functional genomics is insertional mutagenesis, such as transfer DNA (T-DNA) tagging, and adapting T-DNA tagging to barnyard millet would enable the systematic characterization of genes, potentially leading to the discovery of traits beneficial for crop improvement[71]. Functional genomics databases creation for barnyard millet could provide comprehensive resources for storing, querying, and analyzing large-scale data sets and these databases integrate various types of data, including metabolite profiles and small RNA (sRNA) data, and offer tools for identifying co-expressed genes and significant biological processes[68,72].
In addition to these specific techniques, by comparing the functional genomics data of barnyard millet across different species and model organisms, researchers can gain insights into conserved genetic mechanisms and evolutionary adaptations[73,74]. Moreover, the integration of bioinformatics and systems biology in barnyard millet research could advance the understanding of its genetic and molecular underpinnings, facilitating the development of improved crop varieties[59,75]. Bioinformatics tools could be used to analyze the barnyard millet genome, identify genes, and uncover genetic variations, while systems biology approaches map out complex interactions between genes, proteins, and metabolites[60,76]. This integration allows for comprehensive analysis of gene expression, regulatory networks, and metabolic pathways[61,77]. By combining these approaches, researchers can better understand the biological processes underlying key traits, such as stress tolerance and nutrient utilization, and apply this knowledge to optimize breeding strategies and develop resilient, high-performing varieties[8,60,74,78].
-
Barnyard millet's potential as a nutritious and climate-adaptable crop is undeniable, but genetic improvement hinges on advancements in breeding techniques. QTL identification and Marker-Assisted Selection offer promising avenues to target desirable traits like yield and disease resistance. Additionally, CRISPR-Cas gene editing holds immense promise for precise improvement, although challenges like incomplete genome knowledge and transformation protocols need to be addressed. By overcoming these limitations and actively pursuing these genetic improvement strategies, researchers can unlock barnyard millet's full potential, significantly contributing to global food security and sustainable agriculture.
-
The authors confirm contribution to the paper as follows: study conception and design: Sahoo JP; data collection: Pradhan PP, Bhuyan P, Nag G; analysis and interpretation of results: Sahoo JP; draft manuscript preparation: Sahoo JP; review & editing: Pradhan PP, Bhuyan P, Nag G; Sahoo JP. All authors reviewed and approved the final version of the manuscript.
-
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
-
The authors declare that they have no conflict of interest.
- Copyright: © 2024 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Pradhan PP, Bhuyan P, Nag G, Sahoo JP. 2024. Genetic improvement of barnyard millet (Echinochloa esculenta): prospective and challenges. Technology in Agronomy 4: e027 doi: 10.48130/tia-0024-0024
Genetic improvement of barnyard millet (Echinochloa esculenta): prospective and challenges
- Received: 23 June 2024
- Revised: 21 July 2024
- Accepted: 07 August 2024
- Published online: 08 October 2024
Abstract: Barnyard millet is a vital yet underutilized cereal crop due to limited genetic research and improvement efforts. This review explores promising avenues like QTL mapping and Marker-Assisted Selection for targeted breeding, along with the exciting possibilities of genomic selection and speed breeding for faster variety development in barnyard millet. Even gene editing with CRISPR-Cas holds promise, but challenges like incomplete genome knowledge need to be addressed. Additionally, bioinformatics and systems biology approaches offer powerful tools to understand the complex interaction of genes and traits in barnyard millet. While limitations exist, actively pursuing these advancements can unlock barnyard millet's full potential, significantly impacting global food security and sustainable agriculture.
-
Key words:
- Barnyard millet /
- Genetic improvement /
- Nutritional quality /
- Stress tolerance /
- Genomic resources












