-
With the global population experiencing rapid growth, the demand for vegetables and oils has steadily increased. Brassicaceae crops play a crucial role in supplying vegetables and oilseeds. However, clubroot disease, caused by the obligate biotrophic protist Plasmodiophora brassicae (P. brassicae), presents a formidable threat to cruciferous crops worldwide. To date, no effective physical or chemical control methods have been established to combat clubroot in Brassicaceae crops. The historical origins of clubroot trace back to its initial identification on the west coast of the Mediterranean Sea in 1737[1]. Since then, this pathogenic agent has spread globally, especially exhibiting a particular affinity for temperate regions and locales characterized by weakly acidic soils[2]. Moreover, the intensified and continuous cultivation practices applied to Brassicaceae crops have accelerated the widespread dissemination of clubroot, transforming it into a profoundly impactful soil-borne disease on a global scale[3]. Clubroot leads to the formation of considerable root galls, which significantly impair plant growth and diminish the yield and quality of economically vital crops such as Chinese cabbage, cabbage, rapeseed, and radish, among others. For instance, in Canada in 2005, canola yield loss reached up to 50%, while in China, Brassicaceae crops annually suffer yield losses of 20%−30%[4,5]. Beyond impacting yields, clubroot substantially influences crop quality, notably causing decreases in seed oil content by 2%−6% in oil-bearing crops[6].
Symptoms of clubroot primarily affect the roots, leading to the development of galls (clubs) on the infected root tissues, which are characterized by abnormal proliferation. These galls severely deplete nutrient levels, disrupt the organization of vascular bundles in the roots, and obstruct the uptake of water and minerals. Consequently, the aboveground portions of the afflicted plants exhibit yellowing, wilting, and eventual demise[7]. In the short term, symptoms in plants cultivated in well-moistened soil may not be immediately apparent, which challenges the identification of P. brassicae root infections[8]. Compared to cabbage, Chinese cabbage, and rapeseed, diagnosing clubroot infection in radish is particularly challenging due to its primary product being the underground taproot. Until now, there has been a lack of effective methods to control clubroot on Brassicaceae crops. As a result, the cultivation of resistant varieties has become the primary approach to proactively prevent the occurrence of clubroot disease.
In recent years, substantial progress in understanding clubroot resistance in Brassicaceae crops has been realized through investigations of pathogen-host dynamics and the identification of pivotal resistance genes. The discovery of clubroot resistance loci, along with the establishment of corresponding molecular markers, has propelled these advancements significantly. This review elucidates the research developments in crucial areas related to P. brassicae, which include its pathogenicity, host resistance and molecular markers, mechanisms of resistance at the molecular level, and the genetic enhancement of resistance. Moreover, the review underscores current challenges and outlines prospective research avenues within the domain of clubroot resistance in Brassicaceae crops.
-
Clubroot in Brassicaceae is caused by infestation with obligate pathogens, specifically P. brassicae. Although the taxonomic status of P. brassicae is still controversial, these pathogens are currently classified within the genus Plasmodiophora, class Phytomyxea, and phylum Cercozoa[9].
Biological characteristics of P. brassicae pathogen
-
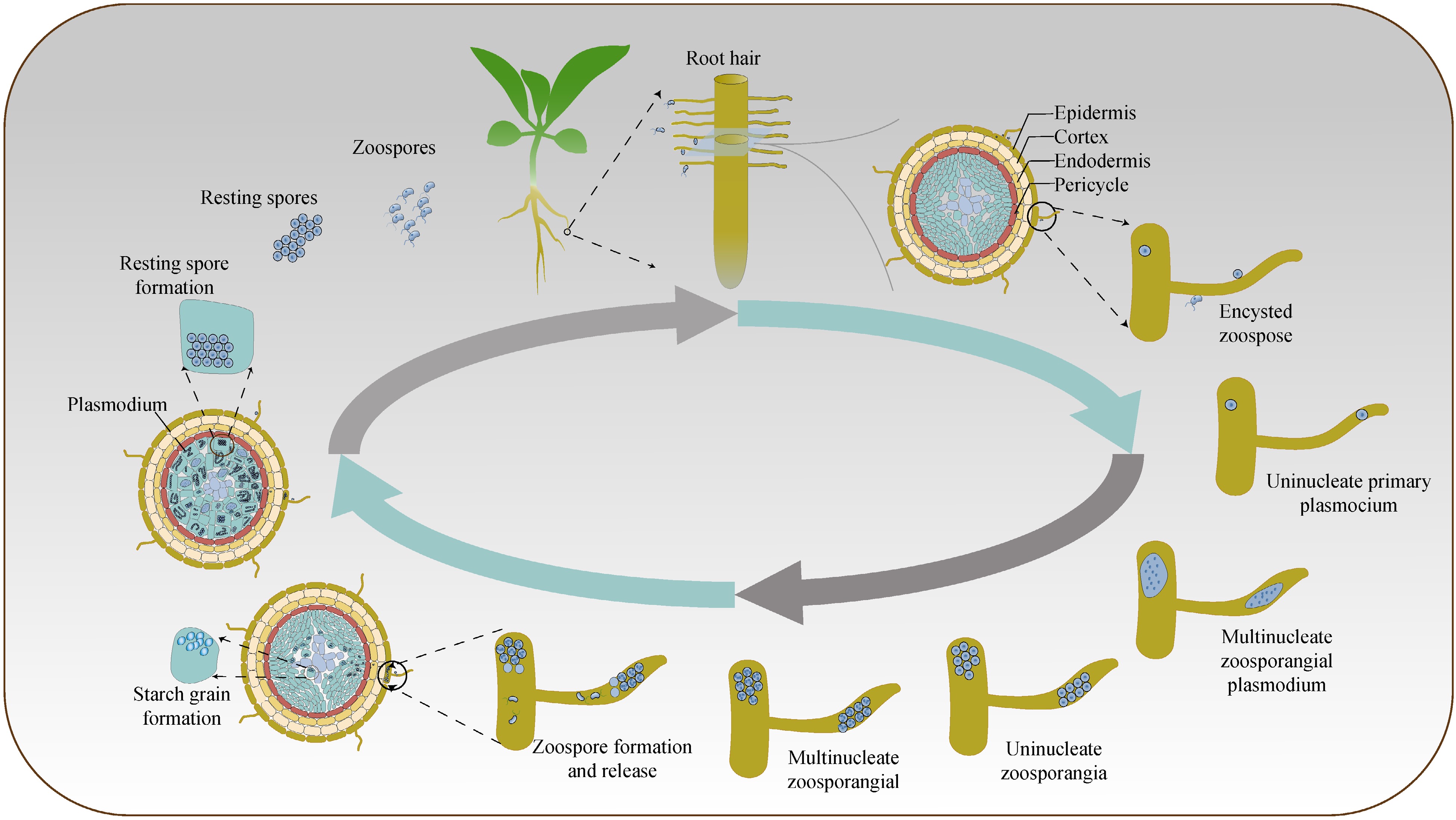
The P. brassicae can infect plants throughout their entire growth period in Brassicaceae. The life cycle of the P. brassicae pathogen is divided into three stages (Fig. 1): dormant spore stage, primary infection (root hair infection) stage, and secondary infection (cortical infection) stage[10,11]. At the dormant spore stage, the pathogen persists in the soil as quiescent spores, which exhibit resilience to adverse environmental conditions and can endure for extended periods. Upon the advent of conducive circumstances, such as the presence of susceptible Brassicaceae plant roots, the dormant spores germinate, transition into the primary infection stage, and discharge motile zoospores. These zoospores navigate through the soil moisture to infect the root hairs of host plants, subsequently penetrating root hair cells and metamorphosing into primary plasmodia. The primary plasmodia grow and multiply within the root hair cells, causing localized swellings and distortion[12]. Progressing to the secondary infection stage, primary plasmodia invade adjacent root cells and traverse the root's cortical tissue, culminating in the genesis of expansive, intricate secondary plasmodia. This results in the formation of secondary plasmodia, which are larger and more complex structures. As the secondary plasmodia develop, they induce the formation of large, multinucleate cells called giant cells within the root cortex. Microscopic examination reveals copious starch granules and zoospores within these cells[13], which fulfill a nutritional role for the pathogen[14].
Primary infection can occur in both host and non-host plants, but secondary infection occurs only in susceptible host plants, except in B. oleracea[12]. The pathogen remains in soil as dormant spores and, upon exposure to root exudates in suitable environments, germinates and liberates primary zoospores. These zoospores migrate toward root hairs using soil moisture and penetrate the cells of the cortex and cambium. Inside the root hairs, the pathogens undergo morphological changes, and the primary protoplasms divide to form primary zoosporangia, which in turn produce secondary zoospores within the primary root cortex. Simultaneously, the pathogen stimulates gene expression related to the biosynthesis of plant hormones such as auxin and cytokinin, transforming the nutrients toward the abnormally enlarged roots and resulting in gall formation. The multinucleated secondary protoplasts formed by the secondary zoospores undergo differentiation into dormant spores, which are subsequently released into the soil upon gall disintegration.
Clubroot is a challenging plant disease to control, mainly reflected in the following points. Firstly, the pathogen P. brassicae has a wide range of hosts, including many non-Brassicaceae species, but symptoms of the disease only manifest in Brassicaceae plants[15]. This broad host range increases the risk of disease spread and diminishes the effectiveness of crop rotation as a management strategy. Secondly, dormant spores could survive in soil free of susceptible host plants for 15 to 20 years[7] and can be easily propagated via contaminated soil, water, equipment, or by humans and fauna. This presents significant hurdles in preventing the introduction and proliferation of the pathogen, especially in regions with intense brassica cultivation or where infected soil has been transported. Thirdly, clubroot disease is influenced by environmental conditions, particularly soil pH and moisture. The primary infection stage is moisture-dependent and prone to disease between 50% and 95% relative humidity. The disease is particularly severe in low-lying and continuously cropped fields[16]. The pathogen thrives in acidic soils with pH levels below 7, and excessive soil moisture or poor drainage can create favorable conditions for its development[17]. Additionally, suitable temperatures, below 10 °C or above 26 °C, are not conducive to the infection of pathogenic bacteria and rapid reproduction. Modifying these factors to suppress disease can be challenging, especially in regions with naturally acidic soils or areas prone to excessive rainfall. Once the soil is polluted, it is not suitable for the cultivation of Brassicaceae plants in the short term[18]. Lastly, no effective chemical pesticides or curative methods are available for infected plants. The complex biology of clubroot, together with its persistence in soil, broad host range, limited resistant cultivars, and environmental predispositions, renders effective control a considerable challenge. Ongoing research, resistance breeding, and integrated management approaches remain imperative for mitigating clubroot's impact on brassica production.
Pathogenesis of P. brassicae
-
The complete pathogenic mechanism of P. brassicae has yet to be completely elucidated. The entire life cycle of P. brassicae involves complex reprogramming of the host, which can be divided into two main aspects: firstly, pathogens interfere with hypocotyl development by preventing cell cycle exit and stimulating cell enlargement through elevated intracellular traffic. In healthy A. thaliana hypocotyls, the progeny cells of the meristems tend to differentiate. However, when stimulated by P. brassicae, infected plants experience enhanced mitotic activity in the meristem regions, leading to a delay in the exit of host hypocotyl cells from the cell cycle. Consequently, the cells maintain a proliferative (mitotic) state, resulting in a significant increase in cell numbers. This shift is accompanied by the production of dormant spores, where local cell expansion takes precedence over host cambium cell proliferation[19,20]. Secondly, during the cell enlargement stage, P. brassica incites endoreduplication by altering host cell cycle regulation, leading to the generation of enlarged cells that house internal spores, providing more space and metabolic capacity to support spore maturation and replication[19,20].
Furthermore, the pathogen evades and suppresses the host defenses while shaping host cell metabolic pathways. Though galls are not vital for the pathogen's life cycle, they are crucial for fostering growth, nutrient uptake, and dormant spore dispersal[21]. By inhibiting xylem formation and augmenting phloem cell numbers, the pathogen P. brassicae can transfer host nutrients to the site of infection (gall). During the early stage of host plant differentiation, pathogen-prompted gene activation associated with phloem differentiation (OPS, BRX, and CVP2) incites root phloem cell proliferation and differentiation, enhancing phloem architecture and enabling galls to outcompete other host organs as a primary carbohydrate repository[22].
Pathotype and identification system of P. brassicae
-
In research, various strains of pathogens are commonly referred to as a 'race'. However, in the natural environment, when P. brassicae infects Brassicaceae crops, it exists in a mixture of races rather than individual isolates[23]. Presently, there's a lack of consensus on the naming convention for different P. brassicae strains[24]. The designation of these strains often relies on the discriminating system utilized in the study, such as the Williams discrimination system, which classifies them as a 'race'.
As an obligate parasite, P. brassicae strains are not amenable to artificial culture or genetic manipulation. As a result, multiple pathogenic types of P. brassicae may be isolated from one source. The resistance of Brassicaceae plants to P. brassicae corresponds to vertical resistance, with specific resistance genes aligning with particular pathogenic types. Since the 19th century, it has been observed that different pathotypes of P. brassicae vary in their pathogenicity towards particular host plants[25]. Inoculation with highly pathogenic strains often yields superior resistance outcomes as compared to a mixture of strains[26]. Therefore, in the breeding of clubroot resistance in Brassicaceae crops, emphasis should be placed on researching the prevalent physiological pathotype in a given region.
Currently, the internationally recognized identification systems for pathotype differentiation of P. brassicae are the Williams Identification System[24] and the European Clubroot Differential (ECD) System[27] (Table 1). The two identification systems consist of 4 and 15 differentiating hosts, respectively. Pathogenic types are determined based on a comprehensive evaluation of host susceptibility within the system. The Williams system is widely utilized in disease resistance breeding due to its limited number of identification hosts, ease of observation, and identification. Among those pathotypes or races, pathotype 4 was widely distributed in China, Japan, and Korea[28,29]. The ECD system, with its array of differential hosts and the inclusion of the single-spore isolation method, allows for more detailed pathogenic type differentiation. Worldwide, at least 24 pathogenic types have been identified using the ECD system. However, the host varieties of the ECD system are mostly European rape, and the substantial workload is not suitable for Brassicaceae crop breeding in Asia. In response to the limitations of these two widely used identification systems, Somé et al. developed another differential set with ten lines of B. napus which classified 20 field collections of P. brassicae into five groups[30]. Furthermore, the Kuginuki system, including 16 hosts from B. rapa (10), B. napus (2), B. oleracea (2), and B. napobrassica (2), was established[31]. These alternatives aim to overcome the constraints inherent to the Williams and ECD systems, providing further options for pathotype identification in Brassicaceae crops. The Canadian clubroot differential (CCD) set, which includes 13 differential hosts, has been used to classify 106 field isolates into 17 distinct pathotypes in Canada[32]. The Sinitic clubroot differential (SCD) set was developed based on 11 differential hosts, and 132 field isolate pathogens were classified into 16 pathotypes[33]. This system is more suitable for the identification of clubroot pathogens in China and provides convenience and support to domestic researchers.
Table 1. Summary for classification systems of P. brassicae pathogens.
Pathotype classification systems Host (cultivar number) Original strain No. Races/pathotypes Ref. Williams' classification system B. oleracea (2) and B. napobrassica (2) − 16 [24] European clubroot differential (ECD) B. rapa (5), B. napus (5), and B. oleracea (5) − 34 [27] Somé system B. napus (10) 20 5 [30] Kuginuki system B. rapa (10), B. napus (2), B. oleracea (2), and B. napobrassica (2) 36 − [31] Canadian clubroot differential (CCD) B. rapa (2), B. napus (9), and B. oleracea (2) 106 17 [32] Sinitic clubroot differential (SCD) B. rapa (7), B. oleracea (2), and B. napobrassica (2) 132 16 [33] Around the world, the predominant pathotypes for clubroot have been identified, disclosing the dominant strains in diverse regions and providing benchmarks for disease prevention. For instance, in China, race 4 is the predominant main pathogenic type[34], while race 8 predominates in Korea[35]. In central Europe, ECDs 16/31/31, races 4, 6, and 7 of Williams system, and P1 and P3 of Somé's system are the most commonly occurring pathogenic types[36]. Moreover, differences in resistance to physiological race 4 from multiple sources have been noted among various Chinese cabbage cultivars. It has been challenging to distinguish the differences in clubroot pathogen sources in Williams' identification system due to the limited number of host varieties[37].
Current research has been seen the advent of several molecular methods to enhance the detection and characterization of P. brassicae pathogenicity. These methods include the use of highly reproducible restriction fragment length polymorphism (RFLP) fingerprinting[37] and random amplified polymorphic DNA (RAPD) molecular markers[38]. In China, race 4 has been further dissected into three pathogenic types based on PCR targeting six genes within the pathogen's genome[39]. Additionally, the integration of single spore isolation, culturing techniques, and genome sequencing can facilitate a more accurate distinction of clubroot races, serving as a dependable foundation for disease resistance breeding in Brassicaceae crops.
-
From the perspective of P. brassicae, the pathogen is an obligate parasite with a life cycle that comprises three stages: dormant spores in the soil, primary infection of root hairs, and secondary infection in the root cortex. Recent studies indicate that P. brassicae's primary infection occurs in both host and non-host species, whereas secondary infection is exclusive to host plants[12,40,41]. Recognition by the host plant during primary infection appears to trigger genetic resistance factors that deter pathogen progression post-primary infection. In the subsequent secondary infection process, the pathogen successfully establishes a pathogen-oriented library, leading to the enlargement of the plant roots and the formation of galls[21,42].
From the host's point of view, the plant innate immune system has two branches: pathogen or microbial-associated molecular patterns (PAMP/MAMP)-triggered immunity (PTI) and effector-triggered immune response (ETI)[43]. PTI immunity triggers plant non-specific defense responses by recognizing pathogen-associated molecular patterns on the surface of pathogenic microorganisms, while ETI immunity uses plant nucleotide-binding site leucine-rich repeats (NB-LRR) to recognize pathogen effectors and trigger specific defense responses.
The physiological resistance mechanisms among host species toward P. brassicae vary. In B. rapa, studies on resistant Chinese cabbage materials have shown that hosts could inhibit the primary stages of pathogens and affect the colonization of P. brassicae[44]. In B. oleracea, both resistant and susceptible materials show primary and secondary infection stages. The main difference between the reactions of resistant and susceptible lines of B. oleracea is the secondary thickening of xylem and no degradation of the cell wall in resistant plants[45].
The development of clubroot is accompanied by the production, transport, and metabolism of various secondary metabolites and plant growth regulators. Particularly, reactions associated with auxin, cytokinin, and brassinosteroid metabolism are common during cell enlargement[46]. As growth regulators play roles in numerous facets of the host-pathogen lifecycle processes, considering their interactions with other growth regulators becomes crucial. Consequently, it is challenging to isolate the effects of individual growth regulators in the interaction between host plants and P. brassicae. Mei et al. discovered that G protein, together with enhanced Ca2+ signal transduction, might promote the production of reactive oxygen species, programmed cell death, and allergic reactions in resistant B. napus after the infection of P. brassicae. Simultaneously, the resistant host can effectively limit gall formation by utilizing the regulatory abilities of auxin and cytokinin[47]. Piao et al. further identified a calcium-sensor gene, BraCBL1.2, highly upregulated during Pb4 infection in CR3-2, implicating its role in the ETI response in B. rapa against P. brassicae[48].
Plant hormonal communication and secondary metabolic pathways are pivotal metrics in host interactions with P. brassicae. A range of plant hormones, including auxin, cytokinin, salicylic acid (SA), jasmonic acid (JA), abscisic acid (ABA), ethylene (ET), and brassinosteroid (BR), in conjunction with metabolites like glucosinolates, flavonoids, polyamines, organic acids, and various enzymes play significant roles in clubroot pathology[49−52]. The ongoing advancements in this field are progressively elucidating their contributions to the pathogenesis and disease resistance framework.
The role of carbohydrates in the interaction between host and P. brassicae
-
Carbohydrates play an important role in root development following clubroot infection[22,53]. Microscopic observation and starch metabolism analysis revealed that P. brassicae could interfere with and hijack the metabolic pathway of Chinese cabbage plants. This interference leads to the transportation of photosynthetic products to the site of root infection, altering the source-sink relationship and facilitating the proliferation of pathogens[13]. The sequencing of P. brassicae has identified genes related to SWEET permease, monosaccharide transporters, and endogenous carbohydrate metabolism. However, the pathogen itself lacks an extracellular invertase gene and heavily relies on exogenous carbohydrates. Sucrose is the main sugar that moves from the source tissue to the sink tissue as the clubroot develops, so the transport of sucrose is crucial for the development of galls[53,54]. To obtain nutrients, the pathogen establishes a long-term feeding relationship with the host and stimulates the phloem-specific expression of sugar transporters AtSWEET11 and AtSWEET12, enabling the transport of sucrose to galls[22]. An increase in the transcription of extracellular sugar-converting enzymes during clubroot infection has also been noted[55]. In infected A. thaliana plants, the transcript levels associated with photosynthesis and carbohydrate transporters are decreased, leading to the downregulation of carbohydrate accumulation due to altered photosynthetic metabolism. This might represent a compensatory mechanism involving the upregulation of the Calvin cycle, tricarboxylic acid cycle, and carbohydrate unloading genes, reflecting the host's effort to curb sugar exploitation by the pathogen[56,57]. Intriguingly, metabolomics analyses have failed to detect sugar components typically associated with clubroot infection in both resistant and susceptible B. napus lines[58].
Trehalose is a non-reducing sugar that can be found in bacteria, fungi, and plants. A metabolome study revealed that the content of trehalose which is typically scarce in higher plants, significantly increases when the plant is infected with P. brassicae. The biosynthesis of trehalose, mediated by the Trehalose-6-phosphate Synthase Gene (PbTPS1), predominantly occurs in plants more susceptible to secondary infections[53,59−61]. This suggests a potential pathogen strategy to manipulate carbohydrate metabolism, converting soluble sugars to starch thereby preserving cell viability[60]. Beyond its role in sugar metabolism and transport, PbTPS1 may contribute to the harmonization of carbohydrate and nitrogen balance within the host[62]. Furthermore, trehalose-6-phosphate serves as a crucial intermediary in the regulation of sucrose synthesis in source organs, influencing sucrose usage in sink organs, and participating in the nocturnal mobilization of sucrose reserves from starch[59].
The role of glucosinolates in the interaction between the host and P. brassicae
-
Brassicaceae is among the few plant families that produce glucosinolates, and P. brassicae is an obligate parasite of Brassicaceae plants, suggesting an association with clubroot. Glucosinolate levels in susceptible Chinese cabbage are markedly elevated in comparison to resistant strains[63]. However, due to the complex composition of glucosinolates, the precise effect of different combinations and doses on hosts remains unclear.
Glucosinolates are categorized into aliphatic, aromatic, and indole groups based on their amino acid precursors[64]. While aliphatic glucosinolates serve as defensive agents against pathogens, aromatic glucosinolates generally correlate with pest defense, and indole glucosinolates, being tied to IAA production, may influence clubroot development directly or indirectly[65,66]. To investigate the role of glucosinolates in clubroot infection, Xu et al. examined the content and composition of glucosinolates in susceptible and resistant Matthiola incana L. at various time points after pathogen inoculation. The data indicated an increase in the levels of three glucosinolates in the roots of the susceptible line, while only aromatic glucosinolates exhibited a significant increase in resistant materials. Additionally, an increase in internal jasmonic acid (JA) concentration was observed in both resistant and susceptible plants. Exogenous JA applications spurred an increase in aliphatic glucosinolates within susceptible lines and amplified aromatic glucosinolates in resistant ones, prompting speculation that the former may expedite the secondary infection, while the latter potentially contributes to defense mechanisms[50]. Glucobrassicin and other indole glucosinolates may support the synthesis of plant auxin, thereby promoting gall growth[67−69].
Wagner et al. conducted a metabolome analysis and discovered the phenyl ethyl glucosinolate (gluconasturtiin) component was higher in susceptible lines compared to resistant lines. Furthermore, by analyzing the metabolite profiles, conducting quantitative pathogen analysis in the DH progeny, and employing quantitative trait loci (QTL) mapping, it was determined that the clubroot resistance allele controlled the increase of glucosinolate[58].
Role of hormones in the interaction between host and P. brassicae
-
Hormones play crucial roles in regulating plant growth, development, and responses to external stresses. Morphological observations have revealed that the formation of root galls in A. thaliana involves initial cortical cell division followed by cell enlargement[70]. It is postulated that the ratio and levels of cytokinin and auxin are key elements in gall tumor development, with cytokinin taking precedence in initial stages and auxin becoming more influential subsequently.
Cytokinin
-
In the initial stages of P. brassicae infection, the observed elevation in active cytokinin levels may be associated with the repression of adenosine kinase 2 (ADK2) activity[42]. RNA sequencing analyses of susceptible plants reveal a pronounced downregulation in cytokinin metabolism, evident during both the incipient and advanced phases of gall formation. Notably, in Arabidopsis clubroot mutants ipt1;3;5;7, where cytokinin synthesis is substantially curtailed, the pathogen's growth is impeded, and its virulence attenuated, indicating cytokinins' pivotal role in the pathogen's lifecycle[71]. Moreover, Bíbová et al. established that countering the auxin-cytokinin ratio with cytokinin antagonist PI-55 in infected Arabidopsis alleviates disease symptoms, highlighting the potential of targeting phytohormonal pathways as a therapeutic avenue[72].
Auxin
-
The accumulation of auxins, particularly indole-3-acetic acid (IAA), is intricately involved in gall tumor progression. A pronounced concentration of IAA in pathogen protoplasts is noted at the gall site during advanced clubroot stages, supporting nutrient assimilation for sporogenous tissue[42,73]. The Gretchen Hagen 3 (GH3) gene, involved in auxin metabolism, exhibits up-regulated transcription levels in plasmodia-containing cells of susceptible plants[46]. Furthermore, genome-wide sequencing of P. brassicae has identified the presence of auxin-responsive PbGH3 protein in the genome[74].
Plant auxin is intricately involved in various aspects of plant life cycles, making it challenging to dissect the specific changes that affect the formation and function of galls. In Brassicaceae plants, multiple pathways related to IAA synthesis are interrelated and may operate simultaneously. One of the key pathways involves the conversion of tryptophan (Trp) into indole-3-acetaldehyde oxime (IAOx) through multiple reactions catalyzed by decarboxylase, followed by the generation of indole-3-methylthiogluconic acid (GSL) through multiple steps. GSL is then converted into indole-3-acetonitrile (IAN) by myrosinase, and finally, IAN is transformed into IAA by nitrilase[75]. P. brassicae may modulate IAA availability by inducing the host's nitrilase genes NIT1 and NIT2, thus influencing auxin balance[70].
Other hormones
-
Salicylic acid (SA) functions as a signal molecule that can activate plant defense responses. Its signal transduction can inhibit the development of clubroot disease during the interaction between the host and P. brassicae[76,77]. PbBSMT, an effector released by P. brassicae into host plants, acts by methylating salicylic acid (SA) and depleting the accumulated SA in the host. This process leads to the suppression of the host's defense response, thereby undermining its ability to defend against the pathogen[78−80]. Additionally, jasmonic acid (JA)-mediated responses counteract P. brassicae infection by enhancing GSL synthesis and nitrilase expression, suggesting an interplay between JA and IAA metabolic pathways[51,63,78,81]. Brassinolide (BR) is associated with cell wall morphology and cell enlargement. Elevated levels of pectin methylesterase (PME) were observed during the early stages of P. brassicae infection. Mutations in the BRI1 gene, which encodes the BR receptor, were found to increase pectin esterification caused by PME overexpression, resulting in reduced cell integrity due to cell wall degradation[42,82]. Schuller et al. also discovered that BR regulates cell expansion in developing galls, and the formation of galls coincides with increased transcription of genes that regulate the BR biosynthesis pathway or encoded positive regulators of BR signaling. Furthermore, BR mutants exhibited impaired formation or maturation of resting spores[46].
Ethylene (ET) is important for the late response of host infection. It modulates the stress response in diseased plants through signal transduction, thereby affecting gall tumor formation[51,83]. Abscisic acid (ABA) is a signal molecule produced by the host to resist drought stress[67]. The content of ABA significantly increases during the late stage of gall formation[84]. Additionally, resistant materials exhibit a higher abundance of proteins associated with ABA metabolism compared to susceptible plants[85].
Other clubroot susceptible/resistant biomarkers
-
Integrating genetic, metabolic, and pathogen resistance strategies, researchers have identified several biochemical markers of clubroot in Brassicaceae plants. The presence of metabolites such as S-methyl cysteine, glutathione, glycine, alanine, and citric acid is positively correlated with susceptibility in B. napus[58,61]. Moreover, the activity of defensive enzymes, such as SOD, POD, and PAL, has been found to reflect the disease resistance of the host. These enzyme activities can serve as useful indices for screening the resistance of B. napus to clubroot.
-
Molecular markers serve as powerful tools for distinguishing genomic sequence variations, facilitating their application in assisted breeding selection, genetic diversity assessment, and gene mapping[86,87]. As genetic and physical map resolutions have improved, the mapping of genes associated with clubroot resistance has become more precise, fostering the identification of various molecular markers related to CR genes (Table 2). Particularly useful are co-dominant markers, which are instrumental in detecting specific resistance genes in plant populations. For example, 'SWU-OA,' a functional marker on chromosome C07, was established through a combination of RNA-Seq and QTL-Seq analyses, yielding substantial insights into the genetics of clubroot resistance in B. oleracea[88]. Zhang et al. performed a genetic assessment on an F2 population resulting from the cross between a resistant turnip and a susceptible Chinese cabbage, leading to the mapping of resistance loci Bcr1 and Bcr2, as well as the formulation of related markers[89]. Inheritance analysis of clubroot resistance in double haploid populations formed by crossing resistant and sensitive lines has led to the development of markers for resistance screening. For example, the marker '09CR.11390652' was developed by analyzing the clubroot resistance inheritance in the double haploid population formed by crossing resistant and susceptible B. rapa lines. This marker accurately distinguishes the Banglim-resistance phenotype within the population[90]. Moreover, the clubroot resistance genes Rcr3 and Rcr9wa have been mapped in B. napus via BSR-Seq analysis. This approach offers a high-density SNP marker array for the delineation of resistance genes and facilitates marker-assisted selection (MAS) strategies[91]. Unraveling the CR genes within breeding materials is of considerable import for the identification of resistant variants and the molecular pyramiding of resistance genes in future breeding efforts. Establishing a correlation between specific CR genes and the respective pathogen types they confer resistance to could greatly propel the development of cultivars with robust and enduring clubroot resistance.
Table 2. CR genes, linked markers, and QTLs in plant species of the Brassicaceae family.
Species CR gene/QTL Linked markers Plant material Mapping population Pathogen race Gene position Ref. B. rapa L. CRa HC352b-SCAR R: DH line 'T136-8' F2 M85 (Race2) A03 [92,100] S: DH line 'Q5' &'K10' CRaim-T R: DH line 'T136-8' S: DH line 'Q5' Crr1 BRMS-088 R: DH line 'G004'
S: DH line 'A9709'F2 Wakayama-01 A08 [101] Crr2 BRMS-096, BRMS-100 R: DH line 'G004'
S: DH line 'A9709'F2 Wakayama-01 A01 [101] Crr3 BrSTS-41, BrSTS-54 R: Inbred line 'NWMR-3 ' F2 Ano-01 A03 [101,102] S: DH line 'A9709 ' Crr4 R: DH line 'G004' F2 Wakayama-01 and Ano-01 A06 [103] S: DH line 'A9709' CRb TRC05, TRC09 R: DH line 'CRShinki' F2 Race 4 A03 [104] S: DH line '94SK' K-3 R: Inbred line 'CCR13685' F2, BC1 Race 4 [105] S: Inbred line 'GHQ11021' TCR79, TCR108 R: DH line 'CRShinki' F2 Pathotype 4 [10] S: Inbred line '702-5' SC2930-Q-FW/SC2930-RV R: DH lines 'T136-8','K13'
S: Inbred line '702-5'F2 M85 (Race2) K04 [106] SC2930-T-FW/SC2930-RV CRc B50-C9-FW/B50-RV R: DH lines 'C9', 'RC22' F2 M85 (Race2) K04 A02 [92,107] B50-6R-FW/B50-RV S: DH line 'Q5' CRk HC688-4-FW/HC688-6-RV A03 [92] HC688-4-FW/HC688-7-RV Crr1a
Crr1bBSA7
BSA2R: DH lines 'G004'
S: DH line 'A9709'F2, BC3F3 Ano-01, Wakayama-01 A08
A08[94] PbBa3.1 sau_um438a R: Turnip 'ECD04'
S: Chinese cabbage 'C59-1'BC1F1 Pb2 A03 [108] PbBa3.3 sau_um398a Pb7 A03 [108] PbBa8.1 cnu_m490a Pb4 A08 [108] Rcr1 (Rpb1) MS7-9 R: Hybrid cv. FN
S: Canola DH line 'ACDC'F1 Leduc-AB-2010 A03 [109] QS_B1.1 BRMS287-aaf SN3523a R: Inbred line 'Siloga'
S: Inbred line 'BJN3'F2 Wakayama-01 A01 [110] QS_B3.3 sau_um028-At4g35530 A03 QS_B8.1 BRPGM0920-BRPGM0173 A08 Rcr4 A03_23710236 R: Canola 'T19'
S: DH line 'ACDC'F2:3, BC1S1 Pathotype 2 A03 [111] Rcr8 A02_18552018 Pathotype 5× A02 Rcr9 A08_10272562 A08 Rcr2 SNP_A03_08 R: Chinese cabbage 'Jazz' BC1S1 Pathotype 3 A03 [112] SNP_A03_09 S: DH line 'DHACDC' A03 CRd yau389, yau376 R: Inbred line '85-74'
S: Inbred line 'BJN3-1'F2:3 Pathotype 2, 4, 7, and 11 A03 [113] CrrA5 TuuYCBRCR404 R: Inbred line '20-2cc1'
S: Inbred line 'ЕС-1'BC1 A05 [114] CRs R: Turnip 'SCNU-T2016'
S: Cabbage 'CC-F920'F2:3 Group 4 A08 [115] Rcr3 A90_A08_SNP_M11 R: '96-6990-2' F1, BC1 Pathotypes 3H and 5X A08 [91] Rcr9wa A90_A08_SNP_M28 S: DH line 'ACDC' A08 BraA3P5G.CRa/bKato1.1 KB59N06, B4732 R: Inbred line 'ECD 02'
S: 'CR 2599', 'CR 1505'F2 Pathotype 2B, 2F, 3A, 3D, 3H, 3O, 5C, 5G, 5I, 5K , 5L, 5X, 6M, 8E, 8J, 8N, and 8P A03 [116] BraA3P5G.CRa/bKato1.2 CRaJY, BGB41 A03 [116] Bcr1 A03-1-192 R: Inbred line '877' F2 A03 [89] Bcr2 A03-1-024 S: Inbred line '255' A08 [89] CRq Br-insert1 R: DH lines 'Y635-10'
S: DH line 'Y177-47'F2 A03 [117] CRA8.1 A08-4346 R: 'H5R' and '409R' F1 PbXm, PbCd, PbZj, PbTc, and PbLx A08 [118] A08-4624 S: 'H5S' and '91-12' CRA3.7 syau-InDel3008 R: Inbred line 'CR510' F2 Pb3 A03 [119] S: Inbred line '59-1' B. oleracea L. CR2a R: 'No. 86-16-5' F2:3 Race 2 LG6 [120] CR2b S: 'CrGC No. 85' LG1 [120] pb-3 4NE11a R: DH line 'Bi' F2:3 ECD16/3/30 LG3 [121] pb-4 2NA8c S: DH line 'Gr' LG1 Pb-Bo1 T2 R: Inbred line 'C10' F2:3 P1 (Ms6 and eH), P2 (K92), P4 (K92-16) and P7 (Pb137-522) LG1 [122] Pb-Bo2 s07.1900 S: DH line 'HDEM' LG2 Pb-Bo3 aa7.1400 LG3 Pb-Bo4 aa9.983 LG4 Pb-Bo5a PBB7b LG5 Pb-Bo5b a18.1400 LG5 Pb-Bo8 c01.980 LG8 Pb-Bo9a aj16.570 LG9 Pb-Bo9b a04.1900 LG9 QTL1 CA69b, CB85a R: Inbred line 'K269'
S: Cabbage line 'Y2A'F2:3 Kamogawa, Anno and Yuki O3 [123] QTL3 CA63 O3 QTL9 CA93 O3 pb-Bo(Anju)1 KBrH059L13 R: DH line 'Anju'
S: DH line 'GC'F2:3 Race 4 O2 [124] pb-Bo(Anju)2 m6R O2 pb-Bo(Anju)3 BRMS-330 O3 pb-Bo(Anju)4 KBrS012D09N1 O7 pb-Bo(GC)1 ACTb, CB10435 O5 CRQTL-GN_1 C2h-1(4), C2h-5(4) R: Inbred line 'C1220'
S: Inbred line 'C1176'F2, F2:3 Race 2, Race 9 O2 [125] CRQTL-GN_2 C3b-3(8), C3a-34(2) O3 CRQTL-YC C3a-65(8) O3 Rcr_C01-1 D134_C01_8,398,944 R: Inbred line 'ECD11'
S: DH line 'T010000DH3'BC1/BC1S1 Pathotype 3A, 2B, 5C, 3D, 5G, 3H, 8J, 5K, 5L and 3O C01 [126] Rcr_C03-1 D134_C03_9,211,088 C03 Rcr_C03-2 D134_C03_585,685 C03 Rcr_C03-3 D134_C03_35,229,606 C03 Rcr_C04-1 D134_C04 _51,280,226 C04 Rcr_C08-1 D134_C08_23,354,593 C08 Rcr_C08-2 D134_C08_28,507,471 C08 B. napus L. Pb-Bn1 OPG03.960 R: DH line 'Darmor-bzh' F1 Pathotypes
4 and 7LG 4 [127] S: Inbred line 'Yudal' PbBn_di_A02 BS008863 Partially resistant 'Aviso' and 'Montego' DH Pathotype P1 A02 [128] PbBn_di_C03 BS006202 C03 PbBn_di_C04 BS007532 C04 PbBn_rsp_C03 BS012716 C03 qCR_A8 Bn-N3-p16098951 R: 'Rutabaga-BF' DH Pathotypes 2, 3, 5, 6 and 8 A08 [129] qCR_A3 UACSSR3667 S: 'UA AlfaGold' A03 Rcr10ECD01 DM_A03_12570715 R: Inbred line 'ECD01' F1, BC1 Pathotypes 3A, 3D, and 3H A03 [130] Rcr9ECD01 DM_A08_10325589 S: Inbred line 'DH16516' A08 ERF034 BnSNP14198336 R: Inbred line 'Kc84R' F2 Pathotypes 2, 4, 7, and 11 A03 [131] S: Inbred line '855S' R. sativus L. Crs1 RSACCCTC4 R: 'Utsugi-gensuke' F2 Ano-01 and Wakayama-01 LG1 [132] REL24, REL6 S: 'Koga benimaru' RsCr1 R09_11227501 R: Inbred line 'BJJ'
S: Inbred line 'XNQ'F2 Pb10 R09 [133] RsCr2 R09_11933628 R09 RsCr3 R09_15947806 R09 RsCr4 R08_16258481 R08 RsCr5 R08_26984449 R08 RsCr6 HB321, HB331 R: Inbred line 'GLX' F2 Pb10 R08 [134] S: Inbred line 'XNQ' B.nigra L. Rcr6 SNP_B03_51, R: 'PI 219576' F1, BC1, F2 Pathotype 3 B07 [135] SNP_B03_52 S: 'CR2748' -
The successful cultivation of disease-resistant cultivars is essential to combat the increasingly severe issue of clubroot disease worldwide. When a singular cultivar possesses multiple resistance (CR) genes that target different pathogenic races, it exhibits robust resistance across a broad range of pathogen variants. Matsumoto et al. found that the combination of CR genes tend to have a synergistic, rather than merely additive, effect on disease resistance[92]. For example, the canola line '618R', which harbors both PbBa8.1 and CRb CR genes, demonstrates strong resistance to field isolates of P. brassicae[77].
Clubroot resistance loci of main Brassicaceae crops
-
Ongoing research, such as quantitative trait locus (QTL) mapping, gene identification, and marker-assisted selection, are pivotal in the breeding of clubroot-resistant Brassicaceae species. These efforts often apply molecular markers to identify disease-resistant materials or employ diverse techniques for generating CR mutants. Following this, resistance genes are pinpointed using QTL mapping, map-based cloning, and homologous cloning of R genes, with a particular focus on elucidating gene function. Research on clubroot has been directed toward major crops within the Brassicaceae family, including Chinese cabbage (B. rapa, AA, 2n = 20), cabbage (B. oleracea, CC, 2n = 18), rapeseed (B. napus, AACC, 2n = 38), among others. To date, at least 33 and 31 CR loci have been identified in B. rapa and B. oleracea, respectively (Table 2). Notably, CR genes such as CRa, CRb, CRc, CRk, Crr1a, Crr1b, Crr2, Crr3, and Crr4 are significant CR genes that have been extensively studied.
At present, the genes CRa[93], Crr1a[94], CRb[95], and CRA3.7.1[96] in B. rapa, along with CRA8.2.4[96] in B. napus, have been subjected to successful functional validation. Both CRa and Crr1a fall into the category of TIR-NBS-LRR resistance genes, with the nucleotide-binding site leucine-rich repeat (NBS-LRR) type being the most ubiquitous category of plant resistance genes. They feature an N-terminal domain akin to that of Drosophila Toll and mammalian interleukin-1 receptors (TIR), which are crucial in signal transduction and pathogen response pathways[97,98]. Furthermore, Wang et al. identified a novel broad-spectrum clubroot resistance gene named WeiTsing (WTS) in Arabidopsis through fine mapping[99]. This gene encodes a small protein that localizes to the endoplasmic reticulum (ER), and its expression in plants induces an immune response. Remarkably, WTS confers resistance to all tested P. brassicae isolates, even those virulent against existing resistant rape cultivars, underscoring its significant potential for use in disease-resistant crop breeding. Additionally, Gravot et al. implemented an integrated methodology combining QTL fine mapping, CRISPR/Cas9-based validation, and in-depth DNA sequence and methylation pattern analyses, discovering that two proximate NLR genes (AT5G47260 and AT5G47280) collaboratively determine a broad-spectrum, quantitative, partial clubroot resistance in Arabidopsis[136].
Research on resistance genes (loci) of clubroot is progressing and deepening, but there are still some controversies. For instance, CRa and CRb genes in B. rapa are thought to be either alleles or closely linked[137]. Then sequence-based physical map to prove that the CRb gene is not an allele of the CRbKato or CRa gene but that they are closely linked[10]. Derived from the Chinese cabbage cultivar 'CR Shinki', the dominant CRb gene was localized to a 64 kb region on chromosome A03 through fine mapping. Sequence analysis of this region identified six tandem coding gene ORFs for NB-LRR, one of which confers clubroot resistance and shares an identical DNA sequence with the CRa gene[95]. Crr1a encodes an NLR protein, and allelic variation analysis of the gene in six Japanese Chinese cabbage cultivars revealed that the Crr1aKinami90_a allele confers resistance, whereas susceptible lines lack a 172-amino acid segment at the allele's C-terminus[138].
Furthermore, it is also noteworthy that the same CR gene or locus may exhibit varying effectiveness against different pathogenic races. In cabbage, the locus CRQTL-YC acts as a major gene against race 2 but only as a minor gene against race 9[125]. Thus, it becomes vital during CR gene research and CR variety breeding to pinpoint the relevant CR gene for the specific pathogenic race used in inoculation.
Homology among clubroot resistance loci
-
Clubroot resistance genes in Brassicaceae species likely originate from comparable ancestral genome regions. A comparison of genetic maps revealed two QTLs (CRQTL-GN_2 and CRQTL-YC) on chromosome C03 of B. oleracea, which had equivalent collinear regions on chromosomes A03, A06, and A08 in B. rapa. Specifically, the clubroot resistance gene C3e-19(14) in chromosome C03 in B. oleracea demonstrates synteny with the Crr1 resistance locus in B. rapa[125]. Moreover, the clubroot resistance loci pb-Bo(Anju)2 and pb-Bo(Anju)4 of B. oleracea were found to be homologous to CRc2 and CRb in B. rapa, respectively. Interestingly, Rcr6 is the clubroot resistance gene identified in the B genome of Brassica plants, which is homologous to chromosome 8 of the A genome[135]. Homology analyses have also indicated that the radish CR locus Crs1 shares sequence similarity with a region on A. thaliana's chromosome 3. Likewise, the radish CR QTL RsCr4 shares homology with the CR QTL PbBa8.1 on B. rapa's chromosome A08 and A. thaliana's chromosome 4[135]. Genome sequencing and BLAST analyses have helped to identify and map the sequence homology between Chinese cabbage and radish genes, establishing a foundation for discovering homologous disease resistance genes in radishes[139]. Additionally, the loci Crr1 (Crr1b), Crr2, CRa, and CRb in B. rapa showed homology with Arabidopsis chromosome 4 regions, while CRk and Crr3 were homologous to Arabidopsis chromosome 3, PbBa3.1 was homologous to Arabidopsis chromosome 5[102,103,108]. Further research utilizing de novo assembly for genome sequencing of ECD04 and comparative genomic analysis identified that 28 published CR loci could be mapped to 15 loci in the ECD04 genome[96]. These insights yield crucial evidence for elucidating the origins and evolutionary trajectory of CR genes within the Brassicaceae family and present a rich genetic repository for advancing disease resistance breeding programs.
Molecular mechanism of Brassicaceae clubroot resistance
-
Plant disease resistance generally manifests as a quantitatively controlled trait involving multiple genes. These diverse genetic backgrounds induce different hormonal changes within host plants, thereby modulating the ability to initiate a defense response to P. brassicae infection[76,140]. In specific interactions, the host's response to pathogen infection is influenced by resistance or susceptibility alleles at different loci.
Metabolomics analysis and pathogen quantitative analysis in segregation progeny can be used to compare and map QTLs involved in resistance and metabolic regulation, providing new insights into molecular mechanisms of clubroot resistance. Metabolic profiling is often employed to compare pairs of plant genotypes that are resistant and susceptible to pathotypes, respectively, so that candidate compounds involved in plant defense or susceptibility can be identified. The use of near-isogenic plant lines can minimize the impact of metabolome analysis results for loci unrelated to resistance or susceptibility plants[141]. For instance, Wagner et al. combined quantitative genetics with metabolomics and pathogen resistance traits in their examination of B. napus's metabolic response to infection. Their findings suggested that different metabolic modules correlate with distinct resistance QTLs, implying participation in individual cellular defense mechanisms[58,81]. Moreover, Hejna et al. used related transcriptomics to analyze the genetic structure of clubroot resistance variation in B. napus inoculated with ECD17/31/31 pathotype. They identified 82 candidate CR genes through multiple analytic approaches, providing a deeper understanding of resistance at the genomic level[142].
In addition, by performing an iTRAQ-based quantitative proteomic study, analyzing the protein expression changes in Chinese cabbage upon P. brassicae infection, the analysis of differentially expressed proteins (DEPs) between resistant and susceptible materials showed that clubroot resistance was associated with the glutathione transferase activity pathway. Additionally, DEPs were significantly enriched in CTK signal transduction or the arginine biosynthesis pathway, both of which are involved in plant defense responses and responses to stimuli[143]. Moon et al. employed proteomic methods to study the interaction between cabbage and P. brassicae, identifying 24 differentially regulated proteins, most of which were involved in oxidative stress, abscisic acid (ABA) metabolism, and glucose-mediated signal transduction pathways[85]. Resistant plants showed an abundance of ABA response proteins and glucose sensor interaction proteins, suggesting their role in the underlying defense framework against P. brassicae. Conversely, susceptible plants expressed higher levels of cobalamin-independent methionine synthase, potentially exacerbating gall tumor development. Genomic and proteomic advancements have significantly accelerated the discovery of CR-related genes and proteins, fundamentally advancing our understanding of clubroot resistance mechanisms. This knowledge base is expected to be transformative for marker-assisted breeding and the enrichment of resistance genes in future crop development.
-
P. brassicae is a tenacious pathogen that inflicts significant damage on Brassicaceae species with persistent effects. Current strategies for clubroot prevention mainly involve agricultural, chemical, and biological methods, along with the selection of resistant varieties. Continuous cropping in the presence of P. brassicae pathogens can lead to increased pathogen populations, and short-term rotations may even facilitate the mutation of the pathogen into more virulent races[144].
In recent years, fungi have been employed as biocontrol agents either through direct application or in combination with seed coating. It has been demonstrated that Acremonium alternatum application before pathogen infection has been shown to slow down the development of P. brassicae and reduce disease severity when applied[145]. The isolate IK726 of Clonostachys rosea inhibited clubroot symptoms, resulting in a 31% and 72% reduction in the disease index in tested winter rapeseed cultivars 'DK Exclaim' and 'DK Platinium', respectively[146]. Additionally, the enzyme chitinase, which degrades chitin plays an important role in bolstering host resistance by obstructing fungal growth[147]. Chitin is a major component of the pathogen's cell wall and chitinase accounts for about a quarter of P. brassicae's cell wall, chitinase activity was enhanced in susceptible and resistant Chinese cabbage lines after inoculation with P. brassicae[148]. Chitin oligosaccharides, degradation products of chitinase, act as elicitors of plant innate immunity, triggering plant defense responses. In the treatment of Chinese cabbage infected with P. brassicae, treating with chitin oligosaccharides for 24 h has been found to reduce pathogen DNA content and alleviate clubroot symptoms[147].
Employing sacrificial bait plants like radish can also deplete the reservoir of resting spores in the soil. Radishes stimulate the germination of latent spores, and their timely removal before secondary infection can significantly diminish soilborne clubroot inoculum[149]. Chemical interventions such as liming have been shown to potentially elevate soil pH, thereby reducing disease severity and increasing yield by up to 98% in susceptible rapeseed variants. While liming and weed control measures may not substantially impact spore densities in plots planted with resistant varieties, combining lime treatments with resistant variants emerges as a robust strategy for clubroot management[150]. Nevertheless, these preventive solutions only mitigate clubroot occurrences and damage, rather than providing a long-term cure. Furthermore, they impose high labor and financial costs and yield only temporary protection[150]. Prolonged or excessive use of liming and chemical agents may lead to detrimental effects on soil quality and groundwater. Consequently, the cultivation of resistant varieties is considered the most cost-effective and efficient clubroot control measure.
Clubroot resistance breeding
-
Molecular-assisted breeding has significantly facilitated the genetic improvement of Brassicaceae clubroot resistance[151,152]. The resistance enhancement process entails the extensive screening of germplasm resources to isolate robustly resistant materials, which are then characterized through molecular markers to identify and introgress resistance genes for novel cultivar development. For instance, Chen et al. adeptly utilized this approach to transfer the clubroot resistance gene from the Chinese cabbage inbred line 'CCR13685' into the inbred line 'GHQ11021'[153].
Gene introgression is a commonly used method for developing resistant rapeseed varieties. Specific disease-resistant rapeseed varieties, notably 'Mendel' and 'Tosca', have been derived through artificial interspecific hybridization and subsequent chromosome doubling techniques pioneered by Diederichse[137,154]. Turnip ECD04, containing 28 known clubroot resistance loci has been widely used as a germplasm for introducing CR genes into Brassicaceae crops[96]. For instance, B. napus 'Mendel', derived from a cross between 'ECD04' and 'ECD15', is extensively cultivated worldwide[155]. Moreover, Shah et al. cultivated a canola line '618R' with dominant resistance by incorporating CR genes PbBa8.1 and CRb[77]. Various radish cultivars, particularly Japanese varieties, are reported to possess strong resistance to P. brassicae, and are also being cultivated as a bait crop[149]. Additionally, amphidiploids resulting from hybridization with Brassica crops exhibit robust clubroot resistance. In previous studies, tetraploid hybridization was employed to produce disease-resistant turnips and cabbage[156]. Through introgression, CR genes from radish have been successfully transferred to Brassica napus-radish chromosome addition lines, locating the significant CR gene against the Ano-01 pathogen strain[127]. By employing classical breeding, transgenic methods, and gene introgression techniques, resistant radish CR genes can be transferred into less resistant species to develop new clubroot-resistant lines. Techniques combining outcrossing and embryo rescue have facilitated the transfer of radish CR genes into B. napus, yielding progeny that lay the foundation for novel resistant rapeseed cultivars[157]. Additionally, resistance material F8-514 was derived from outcrosses (B. oleracea var. alboglabra Y101 × B. napus) using embryo rescue, backcrossing, resistance identification, and marker-assisted selection[158]. Furthermore, Zhu et al. demonstrated successful introgression of three CR genes (CRa, CRb, and Pb8.1) from B. rapa into B. oleracea[159], thus providing essential germplasm for clubroot resistance breeding and capitalizing on heterosis in vegetable crops. Specifically, the refined CRISPR/Cas9-based cisgenic vector system offers a rapid breeding pathway, enabling the development of canola germplasms with marker-free selection and stable clubroot resistance conferred by Rcr1 within a two-year timeframe[160].
-
The obligate parasitic nature of P. brassicae precludes its artificial culture, often resulting in a mosaic of pathogenic forms and potentially compromising pathotype identification accuracy. The pathogen is capable of mutation during its lifecycle, jeopardizing the durability of resistance in host varieties and, in certain cases, reverting them to a susceptible state. The limitations inherent to classical typing systems, such as the Williams or ECDs methodologies, underscore the need for improved identification frameworks that can accurately integrate host-pathogen interactions. In the future, it is imperative to establish more accurate, convenient, and widely applicable identification systems based on the combinations of host cultivars, pathogen types, and corresponding resistance identification results. Moreover, the application of single-cell separation techniques coupled with whole-genome resequencing promises to distinguish pathotypic sequence variations and foster the development of corresponding molecular markers. In the future, high-generation inbred lines or DH lines containing molecular markers of clubroot resistance genes can be utilized to identify physiological pathogen races. This scientific approach will aid in the finer classification and comprehension of P. brassicae pathotypic dynamics.
Pathogenicity, host resistance, and pathogen-host interaction of clubroot disease
-
The pathogenic mechanism of P. brassicae and the response of host plants to its infection are highly complex, involving the production, transportation, and metabolism of various substances. Plant hormones, secondary metabolites, and associated proteins are implicated in clubroot disease, but their precise roles in pathogen-host interactions have not been fully elucidated. As research progresses, the roles played by these substances in pathogen or host disease resistance are gradually being discovered, which is crucial for the control and prevention of clubroot disease.
Recently, with the availability of a genome database for P. brassicae and important Brassicaceae plants (e.g., the model plant A. thaliana), mutants of reported resistance-related genes have been used to study the differential expression of proteins/genes between resistant and susceptible plant lines. The identification of functional genes related to resistance or susceptibility will provide a foundation for understanding the molecular mechanisms involved in pathogen-host interactions.
Genetic improvement of resistance
-
The genetic intricacies underlying clubroot resistance pose significant challenges to breeding fully resistant pure lines. The variability of parental genetic backgrounds, pathogen strains, and the plethora of unreported molecular markers cloud the understanding of resistance gene interactions. Consequently, there are currently limited disease-resistant varieties available for production. Although some molecular markers derived from disease-resistance genes have been utilized for marker-assisted breeding, certain markers still lack polymorphism between susceptible and resistant plant lines. Therefore, the breeding of resistant varieties necessitates the development of more co-dominant markers and new markers for different resistance loci to enhance resistance screening.
The judicious choice of disease-resistant cultivars represents the most economical and effective strategy for clubroot management. However, resistance performance against the spectrum of P. brassicae pathotypes is inconsistent, and cultivars with specificity for a single pathotype may falter against the pathogen's proclivity for rapid evolution. Thus, developing cultivars endowed with a robust array of CR genes presents an imperative yet formidable objective to enhance resistance durability and breadth across cultivars. Leveraging state-of-the-art genomic techniques and high-throughput sequencing to complement traditional breeding will propel the identification of universally resistant genotypes, the concentration of resistance genes, and the realization of cultivars endowed with enduring and broad-spectrum resistance.
Integrated disease control strategy
-
The current strategy for comprehensive control and prevention of clubroot involves breeding and application of resistant cultivars, along with the implementation of rational farming practices and the use of chemical and biological controls. The exploration and exploitation of potent antagonistic microbiota and endophytic fungi capable of mitigating clubroot will create avenues for reducing the disease burden, thus harmonizing the objectives of economic viability and ecological stewardship.
In summary, within the contemporary scientific research landscape, experts are dedicating substantial efforts to conducting an in-depth analysis of the etiology of clubroot disease. The goal is to decipher the complex infection mechanisms and develop a suite of both stable and effective control strategies. With the continuous evolution of technology, the implementation of innovative techniques is anticipated to markedly improve our understanding of the biological traits of clubroot disease, thereby offering significant support for its successful management and treatment.
-
The authors confirm contribution to the paper as follows: study conception and revising the manuscript: Liu L; writing the draft manuscript: Ma Y, Meng Y; reviewing and creating figures and tables: Wang Y, Xu L, Zhang X, Wang L, Li B; reviewing and improving the manuscript: Chen Y, Yuan Y, Wei X, Cui F. All authors reviewed the results and approved the final version of the manuscript.
-
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
This work was supported by grants from the Natural Science Foundation of Jiangsu Province (BK20220573), Key Technology R & D Program of Jiangsu Province (BE2023366), Seed Industry Revitalization Project (Grant No. JBGS(2021)071), the earmarked fund for Jiangsu Agricultural Industry Technology System (JATS (2023) 421) and the Project Founded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
-
The authors declare that they have no conflict of interest.
-
# Authors contributed equally: Yinbo Ma, Yue Meng, Yan Wang
- Copyright: © 2024 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Ma Y, Meng Y, Wang Y, Xu L, Chen Y, et al. 2024. Research progress on clubroot disease in Brassicaceae crops – advances and perspectives. Vegetable Research 4: e022 doi: 10.48130/vegres-0024-0021
Research progress on clubroot disease in Brassicaceae crops – advances and perspectives
- Received: 01 January 2024
- Revised: 17 May 2024
- Accepted: 06 June 2024
- Published online: 16 July 2024
Abstract: Clubroot is a significant soil-borne disease that poses a severe threat to Brassicaceae crops, such as Chinese cabbage, cabbage, rapeseed, cauliflower, broccoli, radish, etc. This disease is caused by an obligate biotrophic protist, Plasmodiophora brassicae Woronin, which induces large root galls that profoundly impair plant growth, yield and quality. The pathogen has a complex life cycle and high genetic diversity, making it challenging to prevent and control. Clubroot poses a serious threat to global Brassicaceae crop production and food security. This review summarizes recent advances in clubroot resistance research, covering aspects of pathogen pathogenicity, host resistance, resistance genes, molecular mechanisms, and genetic improvement strategies. It also identifies current clubroot challenges and suggests future directions for better understanding pathogen-host interactions, developing more durable and broad-spectrum resistance, and implementing integrated disease management practices. This review aims to provide useful insights and recommendations for the effective prevention and control of clubroot disease, promoting the sustainable and healthy development of the Brassicaceae crop industry.
-
Key words:
- Clubroot disease /
- Brassica crops /
- Pathogenicity /
- Resistance markers /
- Resistance improvement