HTML
-
The genus Talaromyces was initially described by Benjamin (1955) to accommodate sexual morph Penicillium species with soft and yellowish ascomata surrounded by multiple layers of interwoven hyphae. Samson et al. (2011) redefined Talaromyces by transferring Penicillium subgenus Biverticillium into Talaromyces regarding the phylogenetic analysis of sequence data from the nuclear ribosomal internal transcribed spacer (ITS) and DNA-dependent RNA polymerase Ⅱ largest subunit (RBP2) genes. Based on a multi-gene phylogeny of a combination of ITS, β-tubulin gene (BenA) and DNA-dependent RNA polymerase Ⅱ second largest subunit (RPB2), Talaromyces has been divided into seven sections, namely as sections Bacillispori, Helici, Islandici, Purpurei, Subinflati, Talaromyces and Trachyspermi (Yilmaz et al. 2014). Currently, phylogenetic analyses of the ITS, BenA, CaM, and RPB2 genes are imperative for new species identification of Talaromyces (Yilmaz et al. 2014, Chen et al. 2016, Houbraken et al. 2020). The number of species in Talaromyces grew rapidly and have now reached more than 170 species (Houbraken et al. 2020).
Species in section Trachyspermi differ from other Talaromyces by conidiophores producing biverticillate phialides and when ascomata produced, have a creamy white or yellow color (Yilmaz et al. 2014, Chen et al. 2016, Wang et al. 2017). Additionally, they grow restrictedly on Czapek yeast autolysate agar (CYA), yeast extract sucrose agar (YES), and dichloran 18% glycerol agar (DG18), and slightly faster on malt extract agar (MEA) (Yilmaz et al. 2014, Chen et al. 2016, Luo et al. 2016, Romero et al. 2016, Wang et al. 2017). Consideration of the phylogeny and morphological features, 27species in Trachyspermi have been accepted by Houbraken et al. (2020) These species are always isolated from a wide range of substrates, including soil, house dust, leaf, wood and fruit from temperate region to tropical area (Chen et al. 2016, Romero et al. 2016, Wang et al. 2016). Their surviving strategy in the low osmotic environment, such as heat and dry-resistance as well, and bioactive compounds were studied comprehensively (Frisvad et al. 2013, Chen et al. 2016, Romero et al. 2016).
Peatlands only cover about 3% of the land surface but currently maintain one-third of global carbon stores (Turetsky et al. 2015). Fungi, with their heterogeneous physiology, metabolic activities, and ecological functions, are recognized as key decomposers of organic matter in these ecosystems (Thormann et al. 2001, Gilbert & Mitchell 2006). Studies of fungi in peatlands mostly focused on the relationship between fungal diversity and environmental factors using sequencing methods (Myers et al. 2012, Asemaninejad et al. 2017). A few culture-dependent studies indicated that Aspergillaceae (accounted for 25-30%) are the most frequently isolated fungi from peatlands (Thormann 2006, Wu et al. 2013), while Talaromyces accounted for 4.2% of the isolates (Wu et al. 2013).
During a survey of fungal diversity in Zoige wetlands, three isolates with biverticillate adpressed penicilli and spheroidal rough walled conidia are isolated from peat soil. They are described here as a new taxon of Talaromyces based on single and combined ITS, BenA, CaM and the RPB2 gene sequences and cultural features on the recommended media.
-
The cultures are isolated by dilution methods from soil samples collected in Zoige wetlands (33°3'54N, 102°34'23.9E) in Qinghai-Tibetan Plateau, China. Dried cultures are deposited in the Herbarium Mycologicum Academiae Sinicae (HMAS), and the ex-type strains are preserved in the China General Microbiological Culture Collection Center (CGMCC, http://www.cgmcc.net/english/OrderingOfCultures.html).
Morphological observations and growth rate
-
Macroscopic characters are studied on CYA, CYA supplemented with 5 % NaCl (CYAS), YES, creatine sucrose agar (CREA), DG18, oatmeal agar (OA) and MEA (Samson et al. 2011). Isolates are inoculated at three points on 90 mm Petri dishes and incubated for 7 d at 25℃ in darkness. Additionally, CYA plates are incubated at 30 and 37℃, and MEA plates were incubated at 30℃. After 7 d of incubation, colony diameters are recorded. The colony texture, degree of sporulation, front and reverse colony colors, the production of soluble pigments and exudates are observed. Acid production on CREA is indicated by a change in the pH sensitive bromocresol purple dye, from a purple to yellow color in media surrounding colonies. Microscope preparations are made from 1-week old colonies grown on MEA (Yilmaz et al. 2014). A Nikon Ellipse 80i light microscope equipped with differential interference contrast (DIC) optics is used to capture digital images.
DNA extraction, PCR amplification, and sequencing
-
Isolates are grown on potato dextrose agar (PDA, Oxoid malt) 1 week, and fungal mycelium was scraped off for genomic DNA extraction. Genomic DNA is extracted by using a simple and rapid "hermolysis" method (Zhang et al. 2010) and stored at -20℃. The ITS, BenA, CaM, and RPB2 genes are amplified and sequenced using methods and primers previously described in Yilmaz et al. (2014).
Phylogenetic analysis
-
The ITS sequences of isolates are firstly blasted on NCBI, resulting in 99% similar to Trachyspermi diversus ex-type strain CBS 320.48, which belongs to Talaromyces section Trachyspermi. Then, sequences of ITS, BenA, CaM, and RPB2 of species belong to section Trachyspermi determined from recent studies (Yilmaz et al. 2014, Chen et al. 2016, Luo et al. 2016, Romero et al. 2016, Wang et al. 2016, 2017, Houbraken et al. 2020), are downloaded from GenBank (Table 1). A single gene data-set is aligned using MAFFT version 7.03 with the Q-INS-I strategy (Katoh & Standley 2013). The ambiguous areas of alignment are located and removed using the Gblocks version 0.91b software program (Castresana 2000). The appropriate nucleotide substitution model for each gene is tested using the Akaike information criterion (AIC) with MrModeltest v2.3 (Nylander 2004). The 'GTRGAMMAI' model is the best model for ITS, BenA and RPB2 sequences, and the'GTRGAMMA' model is the best model for CaM sequences. The model for multi-gene analysis is the combination of all models of the single gene. Talaromyces purpurogenus in Talaromyces section Talaromyces is set as outgroup in each analysis.
Table 1. Species used in phylogenetic analyses
Species name Collection numbera Accession number BenA CaM ITS RPB2 T. aerius CBS 140611T KU866835 KU866731 KU866647 KU866991 T. albobiverticillius CBS 133440T KF114778 KJ885258 HQ605705 KM023310 T. albobiverticillius CBS 133441 KF114777 - - - T. assiutensis CBS 147.78T KJ865720 KJ885260 JN899323 KM023305 T. assiutensis CBS 645.80 KF114802 - JN899334 - T. assiutensis CBS 116554 KM066124 - KM066167 T. atroroseus CBS 133442T KF114789 KJ775418 KF114747 KM023288 T. atroroseus DTO 270-D5 KJ775227 - KJ775734 - T. atroroseus DTO 270-D6 KJ775228 - KJ775735 - T. austrocalifornicus CBS 644.95T KJ865732 KJ885261 JN899357 - T. convolutus CBS 100537T KF114773 - JN899330 JN121414 T. diversus CBS 320.48T KJ865723 KJ885268 KJ865740 KM023285 T. diversus DTO 131-I6 KJ775193 - KJ775700 - T. diversus DTO 133-A7 KJ775194 - KJ775701 - T. erythromellis CBS 644.80T HQ156945 KJ885270 JN899383 KM023290 T. heiheensis HMAS 248789T KX447525 KX447532 KX447526 KX447529 T. minioluteus CBS 642.68T KF114799 KJ885273 JN899346 JF417443 T. minioluteus CBS 137.84 KF114798 - KM066171 - T. minioluteus CBS 270.35 KM066129 - KM066172 - T. peaticola CGMCC 3.18620 T MF284705 MF284703 MF135613 MF284704 T. peaticola CGMCC3.18767 MF960859 MF960861 MF960857 MF960863 T. peaticola CGMCC3.18768 MF960860 MF960862 MF960858 MF960864 T. purpurogenus CBS 286.36T JX315639 KF741947 JN899372 JX315709 T. rubrifaciens CGMCC 3.17658T KR855649 KR855653 KR855658 KR855663 T. solicola CBS 133445T GU385731 KJ885279 FJ160264 KM023295 T. solicola CBS 133446 KF114775 - KF114730 - T. systylus BAFCcult 3419T KR233838 KR233837 KP026917 - T. trachyspermus CBS 118438 KM066128 - KM066166 - T. trachyspermus CBS 116556 KM066126 - KM066170 - T. trachyspermus CBS 373.48T KF114803 KJ885281 JN899354 JF417432 T. ucrainicus CBS 162.67T KF114771 KJ885282 JN899394 KM023289 T. ucrainicus CBS 127.64 - - KM066173 - T. ucrainicus CBS 583.72A KM066130 - KM066174 - T. udagawae CBS 579.72T KF114796 KX961260 JN899350 - Combined sequences of the ITS, BenA, CaM, and RPB2 are concatenated using the Sequence Matrix for Windows version 1.7.8 (Vaidya et al. 2011), and missing data are treated as gaps. Single and combined genes are analyzed using maximum likelihood (ML) performed in RAxML (Stamatakis 2006) implemented in raxmlGUI v.1.3 (Silvestro & Michalak 2012) with rapid bootstrap analysis with 1000 replicates. For Bayesian analyses, the posterior probabilities were determined by Markov chain Monte Carlo sampling (MCMC) in MrBayes v3.2 (Ronquist et al. 2012) based on the best models from MrModeltest. An average standard deviation of < 0.01 for split frequencies is used to suggest a convergence between parallel runs. The first 25% of sampled trees were discarded as 'urn-in' Trees are figured in FigTree v1.4.2 (Rambaut 2014). Bootstrap values higher than 70% from Paup (BSMP), RAxML (BSML), and Bayesian posterior probabilities (BYPP) greater than 0.95 are indicated in the phylogenetic trees.
Soil collections and fungal isolation
-
A first phylogeny concerning all currently accepted species in section Trachyspermi, including the type stain of our new isolates, was performed by using a sequence data-set of combined ITS (448 bp), BenA (351 bp), CaM (476 bp), and RPB2 (841 bp) genes (Fig. 1). The phylogenetic tree presented our putative new species Talaromyces peaticola (CGMCC 3.18620) positioned robustly in section Trachysperm. Within section, T. peaticola (CGMCC 3.18620), Talaromyces clemensii (PPRI 26753), and T. diversus (CBS 320.48) formed a clade with strong support (100, BSMP / 100, BSML / 1.00, BYPP). Within this clade, CGMCC 3.18620 and T. diversus (CBS 320.48) formed a distinct subclade with strong support (99, BSMP / 100, BSML / 1.00, BYPP) as well, suggesting CGMCC 3.18620 is closely related to T. diversus. Consideration of the limited resolution of the ITS in the Trichocomaceae, BenA was proposed as the secondary DNA barcode for Talaromyces (Yilmaz et al. 2014). A phylogenetic tree was reconstructed by employing all available sequences BenA of species in section Trachysperm (Fig. 2). It presented that all isolates of T. peaticola and T. diversus formed a clade with strong support (100, BSMP / 100, BSML / 1.00, BYPP). Within this clade, T. peaticola formed a distinct subclade with strong support (95, BSMP / 75, BSML / 1.00, BYPP) separated from T. diversus, suggesting T. diversus is new taxon.

Figure 1. Phylogenetic tree generated from ML analysis combined ITS, BenA, CAM and RPB2 sequence data for Talaromyces section Trachyspermi. Talaromyces purpurogenus was chosen as outgroup. Bootstrap values higher than 70% for MP analysis (BSMP) (left) and ML analysis (BSML) (middle) are given above the nodes respectively. Bayesian posterior probabilities greater than 0.95 are indicated (BYPP) (right). * indicates bootstrap values of less than 70% or Bayesian posterior probabilities lower than 0.95 for a lineage. T indicates the ex-living type.

Figure 2. Phylogenetic tree generated from ML analysis BenA sequence data for Talaromyces section Trachyspermi. Talaromyces purpurogenus was chosen as an outgroup. Bootstrap values higher than 70% for MP analysis (BSMP) (left) and ML analysis (BSML) (middle) are given above the nodes respectively. Bayesian posterior probabilities greater than 0.95 are indicated (BYPP) (right). * indicates bootstrap values of less than 70% or Bayesian posterior probabilities lower than 0.95 for a lineage. T indicates the ex-living type.
Talaromyces peaticola Jian Q. Tian & Jing Z. Sun sp. nov. Fig. 3
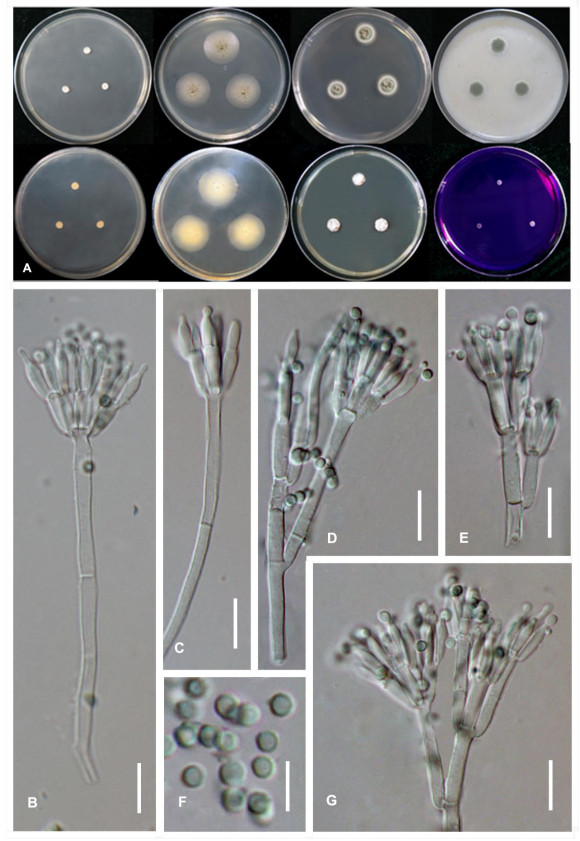
Figure 3. Morphological characters of T. peaticola (ex-type CGMCC3.18620). A Colonies from left to right (top row) CYA, MEA, YES, and OA; (bottom row) CYA reverse, MEA reverse, DG 18 and CREA. B–G Conidiophores. H Conidia. Scale bars: B–C = 20 μm, D–H = 10 μm.
Index Fungorum number: IF553909; Facesoffungi number: FoF 09706
Etymology-peaticola, the stem peati- refers to the substrate that the type strain is isolated, the ending -cola means"weller, inhabit".
Diagnosis-Colonies slow-growing and concave in centers on CYA at 25℃, extremely slow-growing on CYA at 30℃ and 37℃, acid production absents on CREA at 25℃; conidiophores biverticillate; conidia globose, smooth-walled.
Colony diam, 7 d, 25℃ (unless stated otherwise): CYA 5.0-6.9 mm; MEA 24.7-24.9 mm; DG18 16.0-16.5 mm; YES 9 -11 mm; CREA 3.3-3.9 mm; OA 11.7-12.5 mm; CYAS 2.9-3.5 mm; CYA 30℃ 4.7-6.6 mm; CYA 37℃ 4.5-4.7 mm; MEA 30 ℃ 35.5-37.8 mm.
Colony characters -CYA 25℃, 7d: colonies slightly raised at center, slightly sulcate; mycelia white; texture velvety; sporulation absent; soluble pigments absent; exudates absent; reverse medium beige (Fig. 3A). MEA 25℃, 7d: colonies low plane; margins low, plane, entire; mycelia white; sporulation loose; conidia grayish green; texture floccose; soluble pigments absent; exudates small clear droplets; reverse bone white (Fig. 3A). YES 25℃, 7d: colonies slightly raised at center, sunken at center, sulcate; margin slow, plane, entire (< 1 mm); mycelia white; texture velvety; sporulation absents to sparse; soluble pigment absent; exudates absent; reverser white (Fig. 3A). DG18 25℃, 7d: colonies raised at center, sunken in the centre, sulcate; margins low, plane, entire (1-2 mm); mycelia white; texture velvety; sporulation moderately dense to dense; conidia dark green; soluble pigments absent; exudate absent (Fig. 3A). OA 25℃, 7d: colonies low, plane; margins low, plane, entire; mycelia white; texture floccose; sporulation dense, conidia dark green; soluble pigments absent; exudates absent (Fig. 3A); CREA 25℃, 7d: restricted growth; acid production absent (Fig. 3A). On MEA 25℃, 7d conidiophores biverticillate (Fig. 3B-G); stipes smooth-walled, 160-200 × 3-4 μm (Fig. 3D, G); metulae, 3-8, divergent, 7.5-11.5 × 2-3 μm; phialides acerose, per metulae, 7.0-13.5×1.5-2.0 μm (Fig. 3E); conidia smooth, in chain, subglobose, globose, smooth-walled, 1.5-2.5×1.5-2.0 μm (Fig. 3H).
Teleomorph-Undetermined
Known distribution-CHINA. Sichuan Province.
Material examined – CHINA. Sichuan Province, Aba Autonomous Prefecture, Hongyuan County, Zoigê wetland, 33o3′54N, 102o34′23.9E, peaty soil, September 15, 2016, Jianqing Tian, ZRT-4 (holotype: HMAS 247296). ex-type living culture: CGMCC3.18620, CGMCC3.18767, CGMCC3.18768. ibid. LB1.17020001.
Notes - Talaromyces peaticola belongs to Talaromyces section Trachyspermi are well supported by the phylogenetic analyses of a combination of ITS, BenA, CaM and RPB2 sequence data-set. BenA marker and multi-gene of ITS, BenA, CaM and RPB2 performed well in differing T. peaticola from T. diversus. It also can be distinguished from other Talaromyces species by colony slow-growing on CYA at 25℃, extremely slow-growing on CYA at 30℃ and 37℃, acid production absents on CREA at 25℃; conidiophores biverticillate; conidia globose, smooth-walled. In spite of T. peaticola phylogenetically and morphologically closed to T. diversus, it could be easily distinguished from T. diversus by the later could not grow on CREA at 25℃ and T. peaticola producing smaller conidia (Yilmaz et al. 2014).
Phylogenetic analyses
-
The taxonomy of Talaromyces was redefined recently on the basis of DNA sequence data, extrolite profiles and other phenotypic features (Yilmaz et al. 2012, 2014, Chen et al. 2016). The phylogenetic analysis resulted from the combined sequence of ITS, BenA, CaM and RPB2 well distinguish Talaromyces peaticola from other Talaromyces species in section Trachyspermi (Fig. 1), which supported combined ITS, BenA, CaM and RPB2 sequence is imperative for new species identification (Yilmaz et al. 2014, Chen et al. 2016). Additionally, phylogenetic analysis conducted by the single gene of BenA well differs T. peaticola from other species within Talaromyces section Trachyspermi, as well as its sister taxon T. diversus, which resulted from a 9 bp difference in BenA locus (448/457). This confirms that BenA, is imperative for new species identification of Talaromyces (Yilmaz et al. 2014, Chen et al. 2016). However, both phylogenetic analyses based on the single gene of ITS and RPB2 not well differ T. peaticola from T. diversus owning the little differences ITS sequence (585/590 bp) and RPB2 sequences (850/852 bp). These results agree with that ITS and RPB2 sequences are insufficiently variable to reliably discriminate species in Talaromyces (Skouboe et al. 1999, Yilmaz et al. 2012, Frisvad et al. 2013, Yilmaz et al. 2014). BenA and CaM sequences perform well in species delimitation of Aspergillaceae, especially in Penicillium, Aspergillus and Talaromyces, and even some intraspecies (Seifert et al. 2007, Visagie et al. 2014, Yilmaz et al. 2014, Chen et al. 2016).
Colony features on seven standardized media and morphological characters on MEA and are recommended as important phenotypic features in the identification of Talaromyces (Visagie et al. 2014, Yilmaz et al. 2014). Talaromyces peaticola characteristically displays restricted growth on CYA and CREA and exudates small clear droplets on MEA (Fig. 3). In spite that T. peaticola has an affinity to T. diversus in all phylogenetic trees (Fig. 1). These two species are readily distinguished from each otherin regard to T. diversus could not grow on CREA at 25℃ and exudates small clear droplets on MEA (Yilmaz et al. 2014). Additionally, the conidia of T. peaticola are smaller than of T. diversus. Therefore, Talaromyces peaticola is introduced as a new taxon.
Zoige peatland is the largest alpine peatland, characterized by high moisture and low temperature. Previous studies have reported that species in genus Talaromyces are one of the major fungal survivals and frequently isolated from peatland (Gilbert & Mitchell 2006, Wu et al. 2013, Grum-Grzhimaylo et al. 2016, Asemaninejad et al. 2017), the ecological function and metabolic activities need to be further explored.
- This research is jointly supported by the National Key Research and Development Program of China (No. 2016YFC0501802) and the Natural Science Foundation of China (no. 31600024).
- Copyright: © 2021 by the author(s). This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
| JQ Tian, YF Wang, JZ Sun. 2021. Talaromyces peaticola (Aspergillaceae, Eurotiales), a new species from the Zoige wetlands, China. Studies in Fungi 6(1):391−400 doi: 10.5943/sif/6/1/29 |












