-
Rice (Oryza sativa L.) is the staple food of more than half of the world’s population, and production is therefore critical to future food security. Research into rice seed biology is therefore attracting increasing attention from both plant scientists and crop breeders, offering the potential to enhance both yield and quality. The structure of mature rice seeds consists of a spikelet hull, seed coat, aleurone layer, endosperm and embryo, with the embryo containing most of the genetic information, and the endosperm storing nutrients and forming the edible component. Rice seeds showing excellent performance during grain development, seed dormancy and germination are critical in guaranteeing high yield and quality. Understanding the regulatory molecular mechanism of key rice seed traits is therefore essential in improving and breeding elite varieties. For example, high-vigor seeds are suitable for direct seeding production, reducing labor costs and increasing planting efficiency[1], while high-yielding, high-quality rice varieties, with emphasis on taste and appearance, are preferred by farmers[2].
In general, rice seed traits are controlled by both endogenous and environmental factors, with phytohormones playing a central role, not only as a critical intrinsic regulator, but also by conveying environmental input. The most well-known plant hormone family includes gibberellin (GA), abscisic acid (ABA), auxin, cytokinin, ethylene, brassinosteroid (BR), jasmonic acid (JA) and strigolactone[3], all of which play essential roles in seed-related traits, such as ripening (ethylene)[4], dormancy (ABA)[5], and germination (GA)[6]. As a major plant growth-promoting phytohormone, BR is widely involved in a number of rice growth and developmental events, including plant growth, leaf angle, tillering, pollen fertility, grain size and seed germination (Fig. 1). Considering the potential application of BR in improving important agronomic traits in rice, this study reviews the role of this plant hormone in seed biology, such as regulation of grain size, grain filling and seed germination[7−9]. The findings provide valuable information for genetic modification of specific hormone pathways aimed at optimizing seed-related traits in rice.
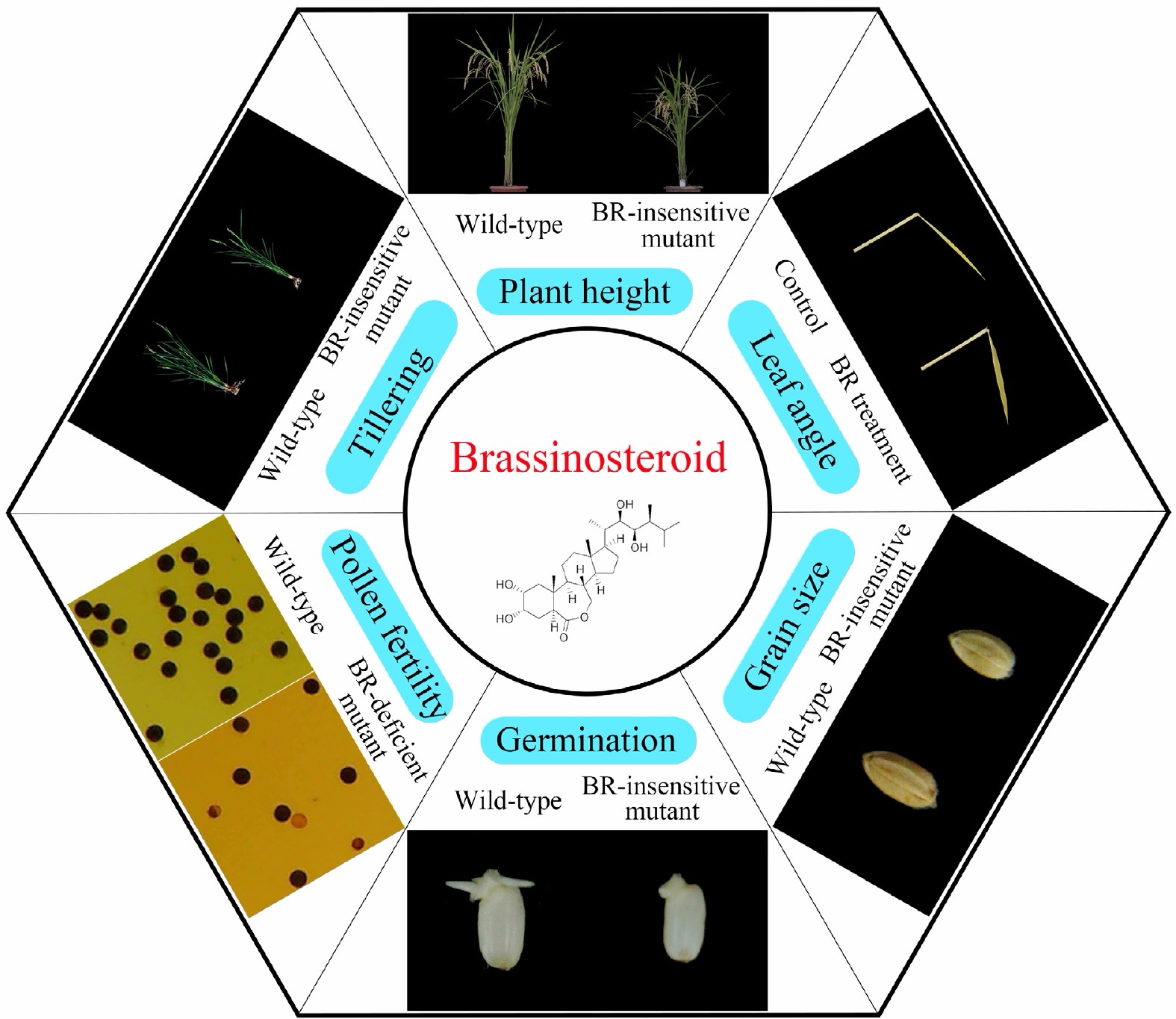
Figure 1.
Brassinosteroid regulates a series of key agronomic traits in rice, including plant height, pollen fertility, leaf angle, tiller number, grain size and seed germination.
The most well-known effect of BR is the promotion of plant growth. BR-deficient rice mutants resulting from mutation of BR biosynthesis genes, such as BRD1[10], D2[11], D11[12], exhibit dwarf and compact phenotypes, while blocking of BR signal transduction was found to trigger a similar phenotype, with reduced plant height and leaf angle[13]. At present, the BR signaling pathway is one of the most understood signal transduction pathways in plants, especially in the model plant Arabidopsis[14]. Although progress in BR-related studies in rice is lagging behind that of Arabidopsis, the primary signaling pathway is conserved. Briefly, BR signals are first sensed by the receptor OsBRI1 and coreceptor OsBAK1, and then transmitted to OsBSK3 kinase and an unidentified phosphatase, thereby suppressing OsGSK2 kinase activity[15,16]. OsGSK2, a GSK3/ SHAGGY-like kinase, is the core negative regulator of the BR signaling pathway, inhibiting BR signals by phosphorylating downstream key transcription factors, such as OsBZR1 and DLT[17−19]. Rice plants overexpressing OsGSK2 or presenting DLT mutation showed similar phenotypes to the BR receptor mutant d61[17,20], highlighting the essential role of BRs in normal growth and plant architecture. In addition, BRs also play multiple roles in rice seed growth and development[15].
-
Grain size is an important agronomic trait closely related to rice yield and quality. As a complex quantitative trait, grain size is controlled by a series of genes, of which more than 80 have so far been cloned[2]. Based on the regulatory pathways, these genes can be divided into six categories: plant hormones, mitogen-activated protein kinase (MAPK) signaling, the G-protein pathway, ubiquitin-proteasome pathway, HAIKU (IKU) pathway and transcriptional regulatory factors[21]. At least four plant hormones; namely, BR, auxin, cytokinin and GA, have been reported to be directly or indirectly involved in the regulation of rice grain size[22], and of these, BR is a major regulator, with more than 20 genes having so far been cloned. According to their specific roles in the BR pathway, these genes can be further divided into three categories: components of BR biosynthesis or signaling, targets of BR signaling pathway, and regulators of the BR pathway (Fig. 2).
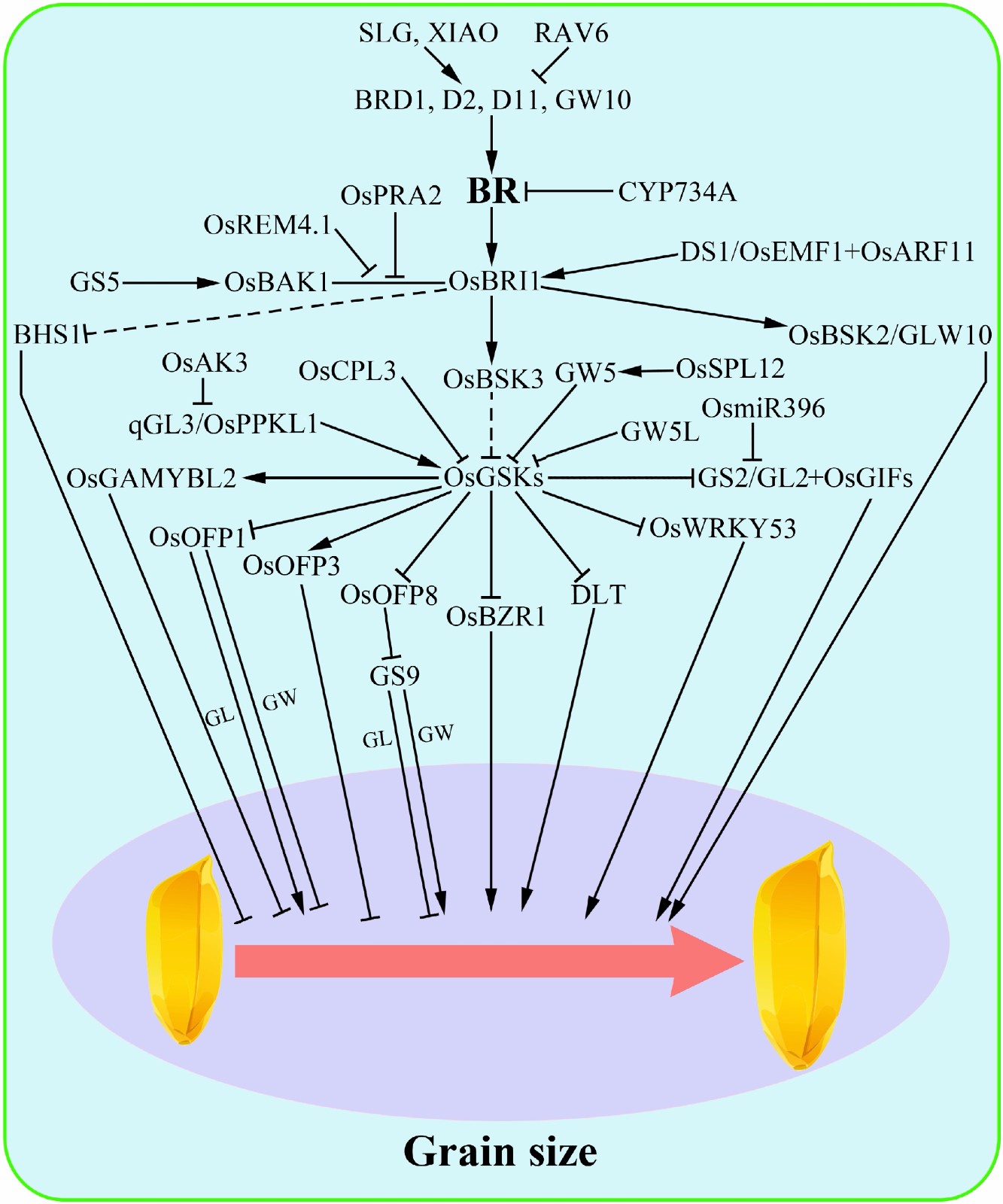
Figure 2.
Brassinosteroid (BR) regulatory networks of grain size in rice. A number of genes related to BR homeostasis and signal transduction play important roles in controlling rice grain size. Notably, GSK protein, which directly interacts with various transcription factors and functional proteins. GL, grain length; GW, grain width. Broken lines indicate indirect or multistep regulation. Arrowheads represent positive regulation.
Components of BR biosynthesis and signaling
-
Mutation of the BR synthesis-related genes BRD1, D2 and D11 produced small grains, suggesting that BR plays positive roles in controlling rice grain size[10−12]. A similar small-grained phenotype was also observed in BR insensitive mutants[7], such as d61 and bzr1[23], while large rice seeds were generated following overexpression of OsBSK3 and knock-down of OsGSK2, respectively[17,24]. Recently, a kinesin-13a protein BHS1 was identified to be a new potential BR signaling component. BHS1 regulates rice grain length by negatively modulating BR signaling downstream of OsBRI1[25]. These results further suggest that BR plays a positive role in regulating rice seed size.
In addition, OsBSK2, a BR-signaling kinase functioning downstream of OsBRI1, positively regulates rice grain size independent of the BR signaling pathway[26]. As the central negative component of the BR signaling pathway, the activity and status of Glycogen synthase kinase 3 (GSK3) family members are also closely related to the regulation of rice grain size. Manipulation of GSK3 family members, such as OsGSK2, OsGSK3 and OsGSK5, was found to alter rice grain size[17,19,27,28]. For example, qGL3 encodes the protein phosphatase OsPPKL1, which contains two Kelch domains, and affects BR signal transduction by dephosphorylating and stabilizing OsGSK3 kinase[27]. Meanwhile, a rare allelic variation of qgl3 in the second Kelch domain was found to result in a long-grained phenotype[29]. Furthermore, OsAK3, which interacts with qGL3 both in vivo and in vitro, results in a longer grain length by controlling cell expansion in rice spikelet glumes[30]. In addition, DLT and BZR1, downstream transcription factors directly regulated by OsGSK2, were found to function as positive regulators of rice grain size[7,17,31]. Recent research further suggests that qGL3 can induce phosphorylation of the 14-3-3 protein OsGF14b, consequently inhibiting OsBZR1 function by promoting cytoplasmic retention and suppressing transcriptional activation activity, which negatively regulated grain length in rice[32]. In addition to its role in the BR signaling pathway, OsPPKL1 is proven to be a cryptic inhibitor of cytokinin phosphorelay which regulates rice grain size, suggesting that PPKL1 may have dual roles in modulating both hormones[33]. Furthermore, disrupting the function of OsCPL3, a member of the RNA Pol II CTD phosphatase-like family, will increase OsGSK2 abundance and decrease OsBZR1 levels, resulting in changes in rice grain size[34].
Targets of the BR signaling pathway
-
In addition to the key elements of BR biosynthesis and signaling, other downstream components of the BR signaling pathway, notably those directly or indirectly controlled by OsGSK2 kinase, also play essential roles in regulating rice seed size. For example, OsGSK2 directly interacts with several members of the OsOFP family proteins, such as OsOFP1, OsOFP3 and OsOFP8, modulating their phosphorylation status, protein stability or activity, and ultimately affecting grain size[35−37]. Interestingly, the activity of phosphorylated OsOFP1 and OsOFP8 was found to decrease[35,37], while the stability and protein accumulation of phosphorylated OsOFP3 increased[36]. In addition, overexpression of OsOFP1 and OsOFP8 significantly increased grain length in rice, while OsOFP3 overexpression had the opposite effect. These results imply that OsGSK2 either enhances the accumulation of negative regulators such as OsOFP3 or suppresses the activity of positive regulators such as OsOFP1 and OsOFP8, thereby acting as a key negative regulator of grain length.
In addition to OsOFPs, several other OsGSK2-interactive proteins related to grain size have also been identified and cloned. For example, OsWRKY53, which is phosphorylated and suppressed by OsGSK2, was found to interact with BZR1 and synergistically control BR-regulated plant traits, including rice grain length[38]. Meanwhile, OsGAMYBL2, another OsGSK2-interacting protein, is destabilized by BR, with up- and down-regulated expression causing a decrease and increase in rice grain size, respectively[39]. Moreover, OsGSK2 was found to directly interact with GS2/GL2/OsGRF4, inhibiting transcription activation activity, and thereby altering cell size[40]. In addition, GS2 has been proven to be involved in the regulation of panicle length, seed shattering and cold resistance in rice[41,42]. As a target of OsmiR396, GS2 also interacts with transcriptional co-activators OsGIFs, forming a complex regulatory module, which predominantly affects cell expansion[43,44].
GS9 is a negative regulator of grain length and a positive regulator of grain width, with gs9 mutant rice seeds exhibiting a slender grain shape. Importantly, GS9 mutation had no effects on rice growth or development, thus improving grain shape without sacrificing yield or other major agronomic traits. Analysis of the underlying molecular mechanism revealed that GS9 directly interacts with OsOFP8 protein, the target of OsGSK2 kinase[45]. Thus, GS9 is indirectly regulated by OsGSK2 via OsOFP8, thereby affecting BR-induced regulation of grain size. Identification of genes downstream of BR that affect grain size would therefore enrich our understanding of the BR signal transduction pathway, and also contribute to the specific improvement of grain size in rice.
Regulators of BR biosynthesis and signaling
-
Some genes control seed size in rice by affecting BR synthesis or signal transduction. For example, RAV6[46], XIAO[47] and SLG[48] regulate rice grain size by modulating the expression of BR biosynthesis genes, such as D2 and D11. In more detail, enhanced expression of SLG improved BR content and increased grain length[48], while knock-out of XIAO led to a distinct BR-deficient phenotype, with short grains[47]. Meanwhile, the effects of RAV6 are more complex. Elevated expression promoted BR content and rice leaf angle, but reduced grain size, possibly due to the complex down-stream target genes of RAV6[46]. CYP734A genes affect the level of bioactive BRs by degrading BRs encoding cytochrome P450 monooxygenases. Plants overexpressing CYP734A4 as a result of a T-DNA insertion showed a typical BR-deficient phenotype, including dwarfing and grain shrinkage[49]. On the other hand, OsREM4.1[50], SG1[51], GS5[52], GW5[53], GW5L[54] and OsSPL12[55] regulate rice grain size by modulating the components of BR signaling. OsREM4.1, encoded by an abscisic acid-induced remorin gene, interacts with OsSERK1/OsBAK1 to inhibit its interaction with OsBRI1[50]. In this way, OsREM4.1 plays a negative role in controlling rice grain size[50]. Similarly, a rice small G protein OsPRA2 also negatively regulates grain size by directly binding to OsBRI1 at the plasma membrane, hence resulting in the dissociation of OsBRI1 from OsBAK1[56]. SG1, which encodes an unknown protein, is a negative regulator of BR signaling and grain size in rice[51], while DS1/OsEMF1 is a positive regulator of rice growth and seed size by interacting with OsARF11 and subsequently activate the expression of OsBRI1[57]. Moreover, enhanced expression of GS5, a serine carboxypeptidase, was found to suppress OsBAK1-7 endocytosis and promote BR signaling, resulting in a larger grain size[52]. Meanwhile, GW5 and its homologous protein GW5L are negative regulators of rice grain width, directly interacting with OsGSK2 to inhibit function[53,54,58]. Furthermore, OsSPL12 is involved in the differentiation of grain size between Indica and Japonica, directly binding to the promoter region of GW5 to regulate transcription. Analysis using the 3000 Rice Genomes Project further indicated interactions between the different alleles of OsSPL12 and GW5, implying that OsSPL12 and GW5 coordinately suppress grain width in Indica[55].
-
Rice germination involves a series of orderly physiological and morphological changes following imbibition and expansion of the seeds, which usually begins with rapid absorption of water and ends with radicle protuberance[59]. Normal seed germination requires a number of prerequisites, mainly light, temperature, water, nutrients and phytohormones[60]. Of the various plant hormones, GA is dominant in promoting seed germination, with GA-deficient mutants in Arabidopsis showing strong dormancy and an abnormal germination phenotype in the absence of exogenous GA treatment[61,62]. In contrast, ABA plays a key role in inducing seed dormancy and inhibiting seed germination[63], with ABA-deficient Arabidopsis mutants showing significantly faster seed germination than the wild-type[64]. Moreover, GA and ABA are antagonistic in regulating seed germination, with spatial and temporal balance determining germination versus dormancy[5]. In addition, GA also promotes seed germination by mobilizing stored starch in rice[65]. This occurs mainly via GA synthesis and secretion into aleurone layer cells in the embryo tissue, which activates the transcription factor GAMYB, thereby initiating the expression of α-amylase and feedback to the endosperm, finally inducing starch hydrolysis into glucose and other small molecular sugars for embryo growth[66]. However, the ABA-inducible OsWRKY51 gene cooperates with the GA-repressible OsWRKY71 gene to inhibit transcriptional activity of GAMYB in rice embryo and aleurone cells[67]. Recently, several genes affecting seed germination and dormancy were cloned in rice mutants, with roles in GA biosynthesis (OsKO1[68]), ABA signal transduction (OsMFT2[69], ISA1[70], OsPP2C09[71]) and ABA metabolism (OsTPP1[72], OsbZIP09[73]). Compared to their respective wild-type controls, these mutants showed either strong germination or dormancy characteristics.
Direct regulation of rice seed germination by BR
-
In addition to GA and ABA, other hormones, including BR, play a role in fine-tuning the rice germination process. For example, shoot length and the germination rate of rice seeds was found to decrease significantly when treated with the BR synthesis inhibitor brassinazole[74]. BR is not only an important regulator of seed germination, but also plays key roles in controlling other agronomic traits related to rice yield, such as grain size, leaf angle, and tiller number. Thus, further understanding of the mechanisms underlying BR-regulated seed germination in rice will aid breeding programs aimed at improving yield-related traits.
Although Arabidopsis and rice share a similar BR signaling pathway, little is known about the mechanism of hypocotyl elongation and germination in rice compared to Arabidopsis[75,76]. In rice, BR-deficient mutants, such as brd1, d2, gns4 and nbg4[10,11,74,77], exhibit a slower germination rate and shorter mesocotyls. Meanwhile, a germination experiment in the dark showed that d61, a BR-insensitive mutant, has a remarkably shorter coleoptile and radicle length[13]. These studies indicate that both BR synthesis and signal transduction are involved in the regulation of rice seed germination and post-germination growth. However, the underlying molecular mechanisms remain largely unknown.
To determine the mechanism of BR-regulated seed germination, a series of BR biosynthesis and signaling related rice mutants were used in a germination assay[78]. The results showed that both the GSK2 overexpressing line and bzr1 mutant germinated slower than their respective wild-type controls. Meanwhile, further experiments revealed that the BZR1-RAmy3D centered transcription module is critical in accelerating the degradation and utilization of starch in both the endosperm and embryo of rice seeds, thus promoting seed germination (Fig. 3). As the key α-amylase gene in rice, RAmy3D, which is also known as αAmy3, acts as a positive regulator of seed germination. Overexpression of αAmy3 could promote rice seed germination under both normal and abiotic stress conditions[79]. Interestingly, RAmy3D is also the target gene in responses to hypoxia and sugar starvation signals, with the regulatory pathway mediated by the CIPK15-SnRK1A-MYBS1-RAmy3D signaling cascade[80] (Fig. 3). More importantly, since RAmy3D is highly expressed during early stages of seed germination, disruption of the BZR1-RAmy3D regulatory module via editing of the BZR1 gene prevented pre-harvest sprouting (PHS) in rice[78]. These findings suggest that the downstream components of the BR signaling pathway could be used to improve the germination and PHS characteristics of rice.
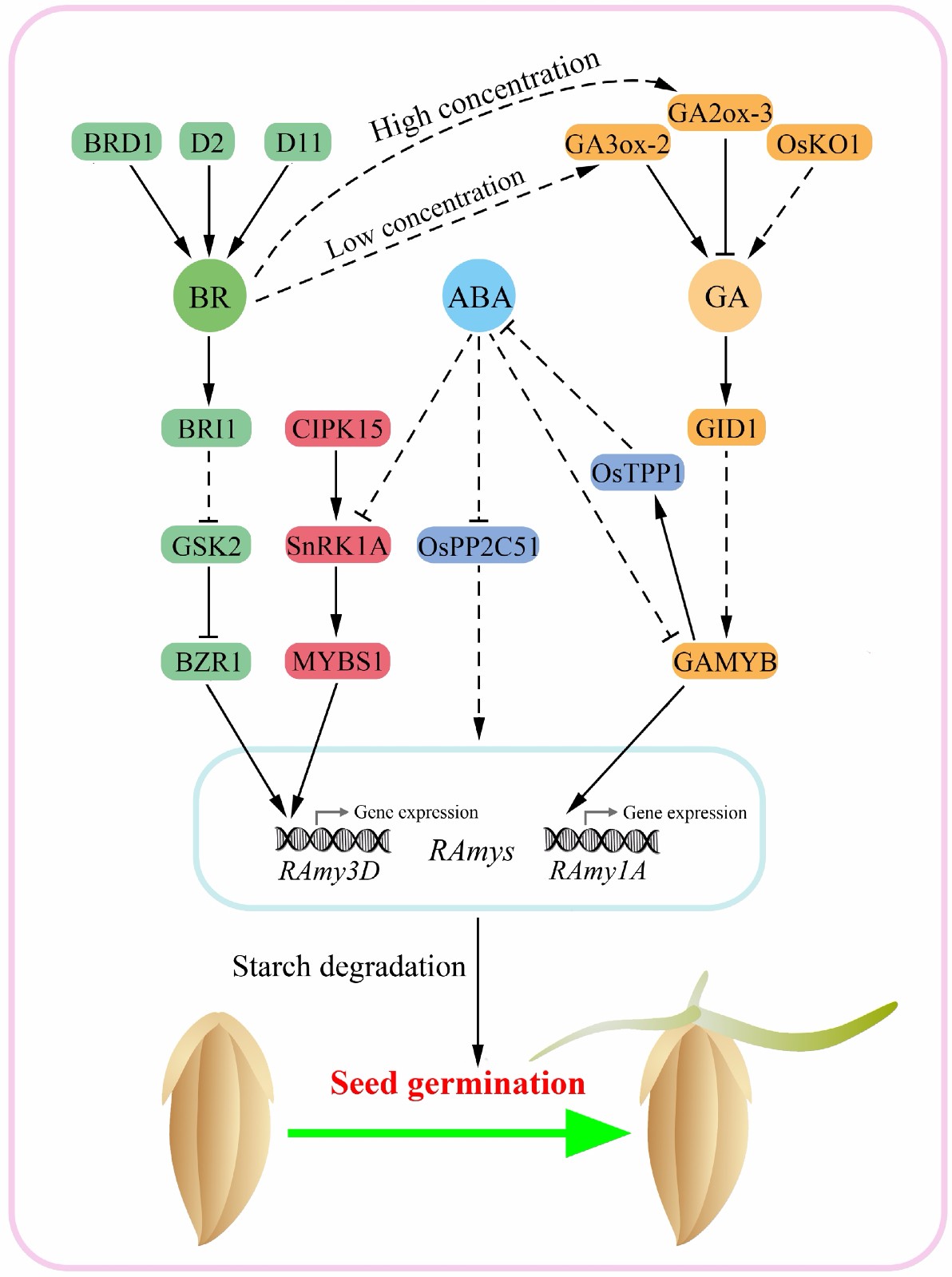
Figure 3.
Molecular regulatory network of brassinosteroid (BR), GA and ABA in co-regulating starch mobilization during rice seed germination. BR regulates the transcription of RAmy3D via the BRI1-GSK2-BZR1 signaling cascade, thereby promoting starch mobilization in the seeds. Different concentrations of BR promote or inhibit GA accumulation by activating the expression of GA3ox-2 and GA2ox-3, respectively. GA promotes starch mobilization through the GAMYB-RAmy1A regulatory module, while ABA interferes with the expression of RAmy1A and RAmy3D by inhibiting the activity of GAMYB in the GA pathway and SnRK1A in the sugar starvation pathway. In addition, ABA down-regulates α-amylase activity through OsPP2C51 via its own signaling pathway. Broken lines indicate indirect or multistep regulation. Arrowheads represent positive regulation.
Co-regulation of rice seed germination via crosstalk between BR and GA
-
Notable progress has been made in determining the molecular mechanisms underlying BR-GA crosstalk during growth regulation in both rice and Arabidopsis[75, 81−84]. Although low levels of BR promoted expression of the GA biosynthesis gene GA3ox-2 in rice, high levels inactivated GA by inducing expression of the GA inactivation gene GA2ox-3, with feedback inhibiting BR biosynthesis[82]. This is also the main reason why a high concentration of BR inhibits rice growth. Based on these studies, the mechanism of BR-GA co-regulated rice seed germination has been gradually revealed. First, GA was found to recover seed germination defects in BR-deficient and insensitive rice seeds, while an iTRAQ proteomic approach identified the differentially abundant target proteins involved in this process[85]. Accordingly, a total of 42 target proteins were identified, five of which are LEA family proteins. Expression of these LEA family members was further found to be suppressed by GA at both the transcript and protein levels. Meanwhile, genetic evidence suggests that LEAs function downstream of both the BR and GA pathways, with LEA mutation inhibiting rice seed germination[85]. Moreover, BR and GA were found to induce the expression of REP-1 in both the rice embryo and endosperm during seed germination, thereby affecting the turnover of storage proteins, including glutelin, which consequently serves to provide more amino acids for embryo growth[74]. The mobilization of starch, which is the most abundant storage component in rice seeds, was also found to be closely related to the control of seed germination. Accordingly, a large amount of transient starch was found to accumulate in the embryos of BR insensitive mutants d61 and bzr1[78], as well as in the GA deficient mutant osko1[68]. BR promotes the degradation of starch through the downstream BZR1-RAmy3D transcription module, which is independent of the GAMYB-RAmy1A transcription module of GA[78]. The interaction node between BR and GA in co-regulation of starch degradation is therefore thought to lie upstream of these transcription factors, thereby influencing hormone biosynthesis. This phenomenon also occurs during the interaction between GA and ABA. For example, GAMYB was found to directly activate expression of OsTPP1, increasing the trehalose content and thereby reducing ABA accumulation in rice seeds[72]. Although the mechanism underlying BR-ABA crosstalk during coordinated seed germination in Arabidopsis and lamina joint inclination in rice have been well established[86−88], direct molecular evidence of the interaction between BR and ABA during co-regulation of seed germination and dormancy has yet to be reported.
-
In addition to grain size and seed germination, BR is also involved in other seed-related traits, such as grain number, grain filling, and starch and protein biosynthesis in the seeds. smg11 is a newly-identified allele of DWARF2 (D2), resulting in small grains and dense panicles, but also an increased number of grains per panicle[89]. GW10, which encodes a P450 subfamily protein, has similar effects on rice grains. Reduced expression of GW10 in the panicles resulted in shorter and narrower rice grains, as well as an increased number of grains per panicle[90]. During the reproductive stage, nutrients are transferred from the leaves to the developing seeds, thereby determining grain filling and subsequent rice yield. Down-regulated expression of OSI-BAK1, a OsBAK1 homologous gene, resulted in a large number of undeveloped green and unfilled grains, with embryonic deletion and developmental delay[91]. Furthermore, ectopic expression of the BR synthetic gene in rice stems, leaves and roots increased both seed number and grain filling, thereby promoting grain yield per plant by about 15%–44%, which suggests that BRs stimulate the flow of assimilates from the source to the seeds[92]. Another study demonstrated that BRs increase the assimilation of glucose to starch biosynthesis in rice, while respective inhibition of D11 and OsBZR1 expression led to defective pollen maturation and reduced starch accumulation in rice seeds[31]. The underlying mechanism involves direct binding of OsBZR1 to the promoter of the CSA gene, promoting expression. CSA, a critical MYB-domain transcription factor, regulates sugar partitioning in rice pollen and seeds by directly regulating the expression of starch synthesis-related genes during seed development[31].
Starch and protein are the two dominant components of the rice endosperm; thus, their constitution and structure determine rice quality[2]. Seed-specific overexpression of the BR biosynthesis gene OsDWF4 in rice was found to alter a variety of seed-related traits, including grain size, chalkiness and the starch structure of endosperm[93]. Furthermore, constitutive expression of ZmD11, an ortholog of rice D11, significantly increased grain size, and the starch and protein content in rice[94]. These results suggest that modulation of BR synthesis and/or signaling affects grain number, grain filling, and the composition and physicochemical properties of rice seeds, thereby altering not only rice yield, but also rice quality traits, such as appearance, nutrition, and eating and cooking indices.
-
BR, as a principal growth-promoting plant hormone, plays a wide range of roles in plant growth and development. In rice, BR has at least three potential applications in breeding practices. First, by attenuating BR biosynthesis or signaling to improve plant architecture. With the reduction in arable land and increasing demands for food, breeding of compact rice varieties complemented by intensive planting has been deemed a practical strategy for reducing competition for water, light and nutrients, thereby improving yield per unit area. Second, BR could also be used to regulate a series of seed-related traits, including grain size, grain filling, grain number and seed components, thereby increasing rice yield and quality. Third, BR involvement in the regulation of seed germination and PHS shows further potential, without affecting the germination rate, with the bzr1 mutant showing slightly delayed germination but strong resistance to PHS[78]. However, due to the pleiotropic effects of BR, improving rice seed-related traits by enhancing the BR pathway also has negative effects on other agronomic traits; for example, increasing the leaf angle and loosening the overall plant architecture. Meanwhile, BR-deficient and insensitive rice mutants typically exhibit dwarfism and a compact plant phenotype, although their grain size is also smaller. At present, the most effective way of applying the positive aspects of BR while avoiding its negative effects is to use weak BR mutants or downstream target genes with specific roles in regulating certain agronomic traits. For example, osdwarf4-1, a weak BR-deficient mutant, causes only limited defects in rice morphology, including a slightly dwarfed stature and more erect leaves, while the yield was higher than that of the wild-type control under dense planting conditions[95]. Another example is GS9, a specific regulator of rice grain size functioning downstream of the BR signaling pathway. GS9 mutation improved rice appearance quality, presenting a slender shape and reduced chalkiness, but without observable negative effects on any other agronomic traits[45].
Rice is one of the most important crops in the world, while its seed traits are directly correlated with yield and quality. Studies aimed at improving seed-related traits are therefore attracting increasing attention from both plant scientists and rice breeders. However, although BR plays comprehensive roles in regulating multiple seed traits, little is known about the underlying molecular mechanism and regulatory network. For example, BR interacts with GA and ABA to co-regulate rice seed germination, as demonstrated by a series of physiological and preliminary molecular studies; however, the exact integrative nodes remain unclear. Future application of powerful and accurate technologies, accompanied by additional genetic resources and a more complete understanding of the BR signaling pathway in rice will therefore help uncover the mechanisms underlying BR-regulated seed traits in rice.
This work was supported by the Hainan Yazhou Bay Seed Lab (B21HJ8105), the National Natural Science Foundation of China (32071984, 31971914), and the Programs from Jiangsu Province Government (BK20200045, JBGS[2021]001, SWYY-154 and PAPD).
-
The authors declare that they have no conflict of interest.
- Copyright: © 2022 by the author(s). Published by Maximum Academic Press on behalf of Hainan Yazhou Bay Seed Laboratory. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Xiong M, Feng G, Gao Q, Zhang C, Li Q, et al. 2022. Brassinosteroid regulation in rice seed biology. Seed Biology 1:2 doi: 10.48130/SeedBio-2022-0002
Brassinosteroid regulation in rice seed biology
- Received: 16 May 2022
- Accepted: 06 July 2022
- Published online: 20 July 2022
Abstract: Rice (Oryza sativa L.) is not only a model monocotyledon plant, but also an important cereal seed crop. Improvements in seed-related traits is the key to obtaining high grain yield and quality, therefore attracting attention from both scientists and crop breeders. In higher plants, brassinosteroid (BR), a major growth-promoting hormone, plays an important role in regulating numerous agronomic traits associated with both vegetative and reproductive growth, thereby presenting huge application potential. Here, we review recent progress into BR regulation in rice seed biology. Both BR biosynthesis and signaling have been shown to regulate grain size, grain filling, grain number, seed germination and biosynthesis of seed components. Thus, considering the pleiotropic effects of BR, strategies aimed at genetic modulation of the BR pathway have been proposed to improve seed-related traits in rice, and therefore, enhance both yield and quality. This review not only strengthens our understanding of the underlying mechanism and regulatory network of BR-regulated key agronomic traits in rice, but also facilitates the future application of BR in rice breeding programs.
-
Key words:
- Overviews /
- Brassinosteroid /
- Regulations /
- Rice /
- Seed /
- Biology













