-
Global climatic changes brought on by both man-made and natural factors have resulted in general rise in temperatures and unpredictable precipitation patterns. This has increased severity and intensity of drought stress with attendant reduction in agricultural productivity. Drought stress is the most damaging abiotic stress, directly threatening food security by limiting crop growth, development, and production. Globally, drought stress has substantially affected production of major food crops causing up to 21%, 40% and more than 50% yield losses in wheat, maize and rice respectively[1]. According to future forecasts, the risk of crop loss could surpass 64%, 68%, and 70% for rice, wheat, and maize, respectively, under increasing dry conditions[2]. A critical solution is to therefore breed drought-tolerant crop varieties that can significantly improve and sustain global crop productivity to feed an ever increasing human population complexed with ongoing climatic changes. Drought tolerant crops such as cassava can sustainably produce high yield under water deficient conditions[3, 4]. Cassava is a perennial tuberous root crop of Amazonian origin[5] with a broad agro-ecological adaptability and inherent tolerance to drought stress[6]. The crop has been classified as a 'drought, war, and famine crop' in the poor world[7] due to its capacity to flourish in low fertility soils and resistance to intermittent and seasonally prolonged drought spells[8]. This has established the crop as a foundation or strategic for food security and poverty alleviation in these regions[7, 9, 10]. Generally plants either resist, avoid, escape, tolerate or recover from drought stress[11]. Plants have embedded these mechanisms at morpho-physiological, biochemical, cellular and molecular levels. Cassava is no exception. The crop deploys multiple drought response mechanisms to maintain growth and yield under water scarce conditions or periods.
Morphological and physiological responses of cassava to drought stress has been widely researched and documented. The crop avoids drought stress by stopping leaf area expansion, reduced transpiration and by its sensitive, fast, and tight stomatal regulation over leaf gas exchange[12, 13] or general reduction in its leaf canopy that decreases transpirational surface area for water conservation[14, 15]. Further response is through decreased stomatal conductance[4], limited leaf formation and extension, increased bud dormancy and extended root growth[16]. All these responses have been directly correlated with changes in biosynthesis, accumulation and distribution of the broad-spectrum phytohormone, abscisic acid (ABA) in most if not all cassava organs and tissues. Indeed cassava varieties have accrued ABA under drought stress[14]. During abiotic stressors such as drought, ABA participates in the coordination of multiple stress signal transduction pathways or the activation of stress sensitive genes in plants. Drought stress activates the transcription factor (TFs) family of genes involved in both ABA-dependent (ABA-D) and ABA-independent (ABA-I) pathways such as the ABA responsive element binding proteins (AREBs/ABFs), Dehydration responsive element binding 2 (DREB2), MYC/MYBs and NAC (NAM, ATAF1, 2 and CUC)[17]. These TFs assist plant in tolerating abiotic stresses and other unfavorable growth conditions, thus making them viable or potential genetic candidates for widespread use in crop breeding or improvement[18]. ABA-D pathways appear to recruit antioxidant and osmoprotectant mechanisms involving glycine betaine, proline, soluble sugars among others compared to ABA-I pathways which generally involves protective proteins[13]. For cassava improvement or breeding objectives, it is important to further investigate and examine the functions of both ABA-D and ABA-I signal transduction pathways and related genes involved in increasing cassava tolerance to drought stress[4, 19−21].
-
Plant growth, development, and responses to both biotic and abiotic stress are all significantly influenced by bZIP TFs[22]. AREBs/ABFs are examples of bZIP TFs that control the expression of genes that are responsive to or are dependent on ABA[23]. By binding to the conserved ABRE cis-elements in the promoter regions, AREB/ABFs exert control over the expression of target genes involved in plant response to abiotic stresses including drought, salinity, heat, oxidative and osmotic[18, 20, 24, 25]. Further, by promoting expression of several late embryogenesis genes, AREBs and ABFs promotes adaptability of different plants or crops to adverse environmental conditions[24]. For example, AREB1/ABF2, AREB2/ABF4 and ABF3 were highly induced by ABA and they both regulated ABRE-dependent ABA signaling in drought stress tolerance in Arabidopsis[25]. Additional candidate AREB/ABFs have also been over-expressed in wheat and rice under drought conditions[26, 27]. In comparison, the functions of bZIP TFs in cassava response to drought have also been studied. For instance Hu et al.[28] revealed that numerous MebZIP genes in the roots and leaves of cassava were activated by drought, indicating their participation in the plant's resistance to drought stress. In this study, eight MebZIP genes (MebZIP41, MebZIP64, MebZIP9, MebZIP58, MebZIP55, MebZIP16, MebZIP72 and MebZIP77) were up-regulated by drought stress while six (MebZIP11, MebZIP27, MebZIP52, MebZIP55, MebZIP64 and MebZIP72) were up-regulated by ABA treatment suggesting their potential role in ABA signaling. Two other MebZIP genes (MebZIP4 and MebZIP52) were over expressed under hydrogen peroxide (H2O2) treatment indicating their potential role in scavenging for reactive oxygen species (ROS) in cassava[28]. Fu et al.[19] reported five other bZIP genes (MebZIP44, MebZIP5, MebZIP53, MebZIP10 and MeHY5/TED5) involved in drought response in cassava.
For ABFs, Feng et al.[20] reported five MeABFs (MeABF1, MeABF2, MeABF3, MeABF4 and MeABF7) that showed significantly higher expression in cassava roots and leaves as a result of drought stress. The MeABFs may activate the MeBADH1 gene by binding to its promoter region, which in turn promotes the production and accumulation of glycine betaine (GB) content in cassava[20]. Under drought, higher GB is synthesized in order to improve tissue water status and insulate biological membranes from ROS, thus osmoprotection[29]. This implied that MeABFs induced the expression of the MeBADH1 gene, which increased GB content that in turn protected the cells from dehydration by preserving osmotic balance and thus improved cassava tolerance to dehydration[20]. Fu et al.[19] also found enhanced expression of ABF2 in cassava subjected to 24-hour PEG-induced dehydration stress, while Orek[30] reported considerable up regulation of ABF2 in all cassava genotypes submitted to water deficit treatment.
NAC transcription factors
-
The NAC TFs are composed of [no apical meristem (NAM)], Arabidopsis transcription activation factor [ATAF1/2] and cup-shaped cotyledon proteins [(CUC2)]. These TFs are involved in general plant growth, development as well as plant adaptability to external stimuli[31]. ABA treatment either activates or suppresses NAC TFs[32] and thus they contribute to drought resistance in plants or crops[33]. For instance, in response to drought stress, NAC genes were activated in rice[34] and wheat[35]. According to Nuruzzaman et al.[36], NAC-mediated stress responses in plants may be directly linked with ROS scavenging and plant leaf senescence. Examples of drought-inducible and ABA-mediated NAC TFs include RD22, RD26/NAC072, ATAF1, and SNAC1[33, 34, 37]. Drought stress and ABA both induced RD22, with its expression considered an ABA early response marker[38, 39]. Nine-cis-epoxycarotenoid dioxygenases (NCED) and ABA-aldehyde oxidase (AAO) are two other NAC TFs involved in ABA biosynthesis pathways[19]. The capacity to withstand drought stress has been associated with NCED3 and RD26 in cotton and transgenic tobacco[40]. RD26/NAC072 facilitates crosstalk between drought and Brassinosteroid signaling and is further involved in ABA- and JA-responsive gene expression[37, 41]. Roles of ATAF1 in drought stress and ABA response have also been reported. For example, ATAF1 positively modulated drought stress in cucumber through ABA-dependent pathway and scavenging for ROS[42]. SNAC1 is also an essential ABA signaling regulator that positively regulates the expression of a variety of ABA signaling genes[34]. Indeed ABA inducible SNAC1 in rice enhanced ABA-induced stomatal closure and improved drought resistance[31].
Similarly, Hu et al.[31] reported expression profiles of cassava NACs TFs (MeNACs) in different cassava genotypes subjected to drought stress. For example MeNAC30 which shares high similarity with ATAF1/ANAC002 that has been shown to be involved in abiotic stress (drought and ABA) responses and leaf senescence[31]. For instance, MeNAC30 that has been reported to have high similarity to ATAF1/ANAC002, was been implicated in responses to ABA, drought and leaf senescence in cassava[31]. Upon re-watering following a drought stress treatment, MeATAF1 displayed varied expression patterns between drought tolerant and drought susceptible cassava accessions[4]. In addition, Orek et al.[4] observed distinct MeSNAC1 expression profiles in drought-tolerant and susceptible cassava cultivars and connected these to stomatal conductance. MeRD22 was also characterized by Lokko et al.[43] as one of the differentially expressed sequence tags (ESTs) with established functions in cassava drought stress responses. Further, a cassava variety that was subjected to PEG-induced dehydration stress, exhibited distinct MeRD22 expression patterns[19]. Utsumi et al.[44] reported up regulation of MeRD26 in diverse cassava genotypes under drought stress using a high-density oligomicroarray analysis.
Diverse cassava genotypes subjected to drought stress treatment showed up-regulation of NCED3, a key gene in ABA biosynthetic pathway which encodes a member of 9-cis-epoxycarotenoid dioxygenases and MeRD26 gene that is involved in the ABA-dependent drought-induced signaling[44]. Both NCED and AAO are key enzymes involved in ABA biosynthetic pathway[19]. Similarly Orek et al.[4] showed that MeNCED3 was up-regulated in different cassava genotypes under drought and linked the gene to an ABA-dependent pathway. Furthermore, MeNCED gene was previously activated in cassava roots and was associated with high ABA levels produced in response to drought stress[45]. Ding et al.[46] reported over-expression of MeRD26 gene in cassava roots and leaves under drought stress. Fujita et al.[33] had also indicated involvement of the MeRD26 gene in drought resistance in cassava based on ABA-dependent stress signaling pathway. Fu et al.[19] showed significant activation of both MeNCED3 and MeRD26 genes in cassava genotypes under polyethylene glycol (PEG)-induced dehydration stress. MeAAO2, MeNCED7 and MeNCED8 genes were also induced in cassava under dehydration[19].
Basic helix-loop-helix (bHLH) transcription factors
-
The bHLH TFs control a range of metabolic and developmental plant processes, including the biosynthesis of secondary metabolites, which is crucial for plant tolerance or adaptation to adverse conditions[47]. For instance, in foxtail millet and rice, bHLHs genes were induced and enhanced these plants’ tolerance to drought stress by activating the ABA and jasmonate signaling pathways, maintaining ROS homeostasis, and triggering stomatal closure[47−49]. Certain bHLH TFs, including MUTE, RD29, EGLE3, GL3, and SPT, have also been linked to processes that improved drought resistance in plants, including stomatal movement, ABA synthesis, extension of root hairs, and the inhibition of leaf growth[50]. The functions of bHLH TFs in the adaptation of cassava to drought stress have also been characterized. As an illustration, the differential expression of two bHLH genes, jasmonic acid ZIM-domain protein 2/9 (JAZ2 and JAZ9), suggested that proteins associated to JA signaling transduction were involved in the response of cassava to under drought conditions[17]. Given that bHLH genes play a role in plant growth and development[51], it has been hypothesized that drought stress may preserve energy by preventing cassava growth so that it can adapt to environmental stresses[17]. Other bHLHs TFs that were identified in cassava under the dehydration stress induced by polyethylene glycol treatment included MeICE1, MebHLH4, MebHLH104, and MebHLH131-like protein[19]. These genes were either up- or down-regulated.
WRKY transcription factors
-
The WRKY TFs are ubiquitous in higher plants and play important roles in a variety of physiological processes and adaptation to adverse environment conditions including leaf senescence and plant response to abiotic stresses such as drought[52, 53]. WRKY TFs are important components of ABA signaling[54], and their over-expression improved drought tolerance in rice[55] and wheat[56]. Differential expression of MeWRKY genes in response to drought stress in various cassava accessions suggested that they contribute to drought stress resistance in cassava via ABA signaling and oxidative stress regulation[57, 58]. Fu et al.[19], for example, found three MeWRKY TFs (WRKY1, WRKY21, and WRKY23) that were variably expressed in response to PEG-induced dehydration stress, as well as consistent ABA-induced expression in cassava roots and leaves. Furthermore, the functional roles of MeWRKY20 and MeWRKY75 in cassava have been analyzed under drought stress[57, 59]. MeWRKY20 and MeWHY1/2/3, for example, reportedly regulated ABA accumulation in cells by inducing the expression of two ABA biosynthetic genes, MeNCED5 and MeNCED1, hence enhancing drought tolerance of wild-type cassava plants via ABA biosynthesis[32, 57, 60]. MeWRKY33, another candidate, also exhibited significant up regulation in drought-treated cassava plants[17]. Wei et al.[58] found that drought stress increased the expression of nine MeWRKY genes in the leaves and roots of different cassava accessions. These included MeWRKY6, MeWRKY11, MeWRKY14, MeWRKY18, MeWRKY20, MeWRKY40, MeWRKY42, MeWRKY49, and MeWRKY83[58]. In conclusion, cassava WRKY genes may play a key role in water intake from soil by roots, resulting in improved drought tolerance[58].
Myeloblastosis (MYB / MYC) transcription factors
-
MYB family proteins serve a variety of roles in plant abiotic stressors such as drought, salt, and cold stress[61, 62]. They have an important role in biosynthesis of secondary metabolites like anthocyanins, flavonols, and lignin[61]. Some MYBs are specifically involved in the regulation of stomatal movement, the control of suberin and cuticular wax production, and the regulation of flower development in response to water stress[63]. Cotton, potato, and Arabidopsis MYB TFs have been shown to be involved in the adaptive response to drought stress[40, 64, 65]. The MYC (RD22BP1/AtMyc2) and MYB (AtMyb2) TFs binds to the cis-elements in the RD22 promoter and activates the RD22 gene[65]. Drought tolerance is increased by ABA-inducible MYB96, which induces stomatal closure via the RD22 gene, up-regulates cuticular wax biosynthetic enzyme genes, and modulates root growth and development[66]. Turyagyenda et al.[21] found a substantial increase in MeMYC2 expression in drought susceptible cassava genotypes compared to non-expression in drought tolerant cassava genotypes under a greenhouse water deficit treatment. In cassava, several other MYBs genes that responded to drought signals have been discovered[19, 62, 67]. For example, Liao et al.[67] found 26 cassava R2R3 MYB family genes that were expressed during water deficiency in cassava. Water deficit treatment resulted in the down regulation of MeMYB2 and MeMYB9 in cassava leaves and up regulation of MeMYB26 in cassava roots[62, 68, 69].
The non-differential expression of MeMYB2 in cassava under PEG-induced dehydration stress suggested that the gene did not play a key part in cassava's ABA-dependent pathway[19]. However, RNAi-mediated MeMYB2 regulation increased drought tolerance in transgenic cassava[62]. Furthermore, drought stress increased the expression of MeMYB21 in cassava[17], whereas Wang et al.[68] identified MeMYB26 as a reliable candidate gene associated with cassava drought tolerance and biomass storage. In response to water deficit treatment, MYB44 and MYB60 gene regulation patterns differed between drought tolerant and drought susceptible cassava genotypes[4, 30]. Cassava exposed to PEG-induced dehydration stress showed varied expression patterns of MeMYB6, MeMYB15, and MeMYB31 in several tissues, including roots[19]. Wang et al.[70] recently found that ABA-induced induction of MeMYB108 with over-expression of gene greatly reduced the rate of drought-induced leaf abscission under drought stress in cassava. As a result, MeMYBs genes may modulate cassava responses to abiotic stress like drought through both ABA-dependent and ABA-independent mechanisms[62]. MeMYBs genes may contribute to drought stress by increasing stomatal closure in the ABA-dependent pathway.
Homeodomain-leucine zipper (HD-Zip) transcription factors
-
The HD-Zip proteins are one of the critical TFs involved in plant growth and development through their regulatory role in the ABA signaling pathway[71]. Arabidopsis, rice, maize, soybean, legume, and banana are examples of crops or plants where HD-Zips have been studied[71]. For example Arabidopsis HD-Zip I subfamily, including AtHB5, AtHB6 and AtHB7 were either up- or down-regulated by drought stress as well as ABA treatment[72, 73]. Other research has also shown that the HD-Zip II and HD-Zip IV subfamilies also respond to drought stress and ABA treatment. Similarly, Ding et al.[71] observed differential expression patterns of multiple MeHDZ I, II, and IV subfamilies in leaves and roots of three different cassava genotypes under drought and PEG treatment. These included nine MeHDZ I subfamily (MeHDZ25, MeHDZ39, MeHDZ38, MeHDZ37, MeHDZ36, MeHDZ23, MeHDZ20, MeHDZ21, and MeHDZ26); three MeHDZ II subfamily (MeHDZ31, MeHDZ32, and MeHDZ34), and four MeHDZ IV subfamily (MeHDZ10, MeHDZ11, MeHDZ15, and MeHDZ55)[71]. Yu et al.[74] previously recorded up-regulation of MeHDS1, a member of the HD-Zip IV subfamily, in cassava during drought stress, with its expression varying more in roots than in leaves. Yu et al.[75] recently found an HD-Zip I TF, MeHDZ14, which was substantially activated by drought stress in several cassava varieties.
TCP transcription factors
-
TEOSINTE BRANCHED 1 (TB1) from maize (Zea mays), CYCLOIDEA (CYC) from snapdragon (Antirrhinum majus), and PROLIFERATING CELL FACTORS 1 and 2 (PCF1 and PCF2) from rice (Oryza sativa) are the first four characterized members of the TCP family of TFs[76]. TCPs regulate a variety of biological processes throughout plant growth and development, including plant architecture, leaf morphogenesis, phytohormone pathways, and response to environmental stimuli[77]. TCP TFs control plant development and defense responses by increasing of bioactive metabolites such as brassinosteroid, jasmonate, and flavonoids[76]. TCP TFs may also play a beneficial regulatory role in plant drought tolerance via an ABA-dependent signaling pathway[78]. Drought tolerance has been improved by over-expression of TCP-TFs in bamboo[78], rice[79], and maize[80]. Similarly, the role of TCP TFs in cassava drought resistance has been studied. For example, Lei et al.[81] discovered 36 non-redundant MeTCPs in drought-stressed cassava seedlings. The drought stress treatment strongly induced seven genes (MeTCP20c, MeTCP20e, MeTCP11a, MeTCP2b, MeTCP19, MeTCP13a, and MeTCP13b)[81]. Furthermore, 22 MeTCPs were highly sensitive to ABA, showing that the MeTCPs genes may be regulated by the ABA signal pathway[81]. Furthermore, MeTCP3a and MeTC4 showed altered expression patterns under drought stress, implying that they may also play vital roles in cassava under abiotic stress conditions[81].
Heat stress transcription factors (HSFs)
-
HSFs play an important role in plant stress response by regulating the expression of stress-responsive genes such as heat shock proteins (Hsps)[82]. Drought and plant hormones like ABA and ethylene have been demonstrated to influence the expression of plant HSF genes. Drought resistance in chickpea (Cicer arietinum L.), for example, was enhanced by over-expression of HSFs such as CarHSFB2[83]. The expression of HSFs genes in cassava has also been studied. Drought stress, for example, increased the transcript levels of MeHsfB3a, MeHsfA6a, MeHsfA2a, and MeHsfA9b, and additional interaction network and co-expression analyses revealed that these HSF genes may interact with Hsp70 family members to withstand environmental stresses in cassava[84]. Zeng et al.[85] discovered several MeHSFs that were up-regulated after treatments with both PEG and ABA, showing that the MeHSFs may play a role in resistance to simulated drought stress via the ABA signaling pathway. HSP90, for example, is critical for drought stress resistance in cassava by regulating ABA and hydrogen peroxide (H2O2). Among cassava's MeHSP90s, MeHSP90.9 transcript was mainly up-regulated during drought stress. MeHSP90.9 may directly activate MeWRKY20 on the W-box element of the MeNCED5 promoter, encoding a major enzyme in ABA biosynthesis and hence regulators of drought stress resistance in cassava[57]. Furthermore, MeHSP90.9 inhibited cassava leaves' ability to accumulate H2O2 during drought stress and positively regulated MeCatalase1 activity, indicating MeHSP90.9 as a possible ROS scavenger.
-
Nuclear Factor Y, Sub-unit A5 (NFYA5) are ubiquitous TFs comprised of three different sub-units (NF-YA, NF-YB, and NF-YC)[86]. NFYA5 is a member of the Arabidopsis NF-YA family whose over-expression promotes ABA-induced stomatal closure, plant survival under drought stress and by positively regulating other drought-responsive genes via the CCAAT box cis-element[86]. Indeed, Arabidopsis plants with NFYA5 over-expression showed significant drought stress resistance[86]. Similarly, in drought-stressed cassava, MeNFYA5 has been implicated in ABA-dependent signaling and stomatal movement[4, 30]. Protein kinase OST1 (open stomata 1) and protein phosphatase ABI1 (ABA insensitive 1) TFs are two critical components of the ABA signaling pathway[87]. OST1 and ABI1 are ABA transduction pathway regulators of Slow Anion Channel-Associated 1 (SLAC1)[87]. ABI1 inhibits ABA signaling, and inhibiting ABI1 could provide a strategy for increasing crop yield under drought stress[88]. Drought/ABA signaling in higher plant guard cells is mediated by the SnRK2 kinase-OST1, which activates the anion channel SLAC1[89]. Active stomatal closure necessitates the SLAC1/OST1 module[89]. OST1 encodes SnRK2, an ABA-activated protein kinase implicated in stomatal closure via ABA stimulation[90]. OST1 and SLAC1 have been associated with limiting water loss thus improving drought tolerance in maize[91]. As part of the drought avoidance strategy, transcriptomic investigation of cassava revealed that acetic acid treatment elevated the expression of ABA signaling-related genes such as MeOST1, MePP2C, and MeTSPO[90]. Drought avoidance in acetic acid-treated cassava plants was enhanced by lower stomatal conductance and transpiration rates, higher leaf relative water content, and higher levels of ABA, chlorophyll, and carotenoid[90]. Suksamran et al.[92] demonstrated that down regulation of MeSLAC1 reduces water loss in cassava during drought stress. Orek et al.[4] reported MeSLAC1 over-expression and down regulation in drought tolerant and drought susceptible cassava genotypes exposed to moisture stress. Ruan et al.[93] observed that drought stress or ABA treatment increased ABI1 expression, whereas Orek et al.[4] reported that elevated levels of MeABI1 in drought tolerant cassava genotypes was consistent with sustained stomatal opening and gradual decrease in stomatal conductance under conditions of water scarcity.
SCaBP5, a Ca2+ binding protein, and PKS3, an interacting protein kinase, act as global modulators of ABA responses[94]. Arabidopsis mutants with suppressed SCaBP5 or PKS3 (scabp5 / pks3) had much lower transpirational water loss, rapid stomatal closure, and improved ABA response in guard cells. SCaBP5 and PKS3 are both involved in a calcium-responsive negative regulatory loop that controls ABA sensitivity[94]. Drought and ABA treatment increase the expression of SCaBP5 and PKS3. Orek et al.[4] showed increased expression of MeSCaBP5 and MePKS3 in cassava genotypes under drought and linked this with a decrease in stomatal conductance, a potential drought avoidance strategy in cassava. Phospholipase D alpha 1 (PLDα1) is a phospholipid hydrolyzing enzyme in plants that plays a role in abiotic stress responses and ABA signaling[95]. PLDα1 mediates ABA modulation of stomatal movement and its increased expression has been observed in response to dehydration and ABA treatment[96]. Elevated expression of PLDα1 improved drought tolerance in Arabidopsis[96] and upland rice[97]. Wang et al.[98] discovered PLDα1 (MesPLDα1-3) in cassava and Orek et al.[4] observed that drought stress increased the expression of MePLDα1 in drought tolerant cassava genotypes compared to its down regulation in drought susceptible genotypes. This corresponded with observed changes in stomatal conductance between the two classes of genotypes[4].
Pyrabactin resistance 1 (PYR1) / Regulatory component of ABA receptor 11 (RCAR 11) act as ABA sensors and regulate protein phosphatase 2Cs (ABI1 and ABI2) via ABA[99]. PYR1 positively regulates ABA-mediated stomatal closure[100]. PYR1 up regulation improved drought tolerance in rice[101] and wheat[102] through positive modulation of ABA signaling. Zhao et al.[103] discovered multiple PYL/R-PP2C-SnRK2 genes that were up regulated in cassava under ABA treatment and abiotic stresses. MePYR1 was found to be involved in cassava signaling or response to ABA[104]. MeCBF3 and MeCBF4, two AP2/EREBP members that were previously linked with low temperature and ABA response, were found to be differently expressed in cassava roots during drought stress[105]. Orek et al.[4] observed increased and decreased expression of MePYR1 in drought tolerant and drought susceptible cassava genotypes respectively under drought stress treatment while Li et al.[17] analyzed a high number of 'response to ABA stimulus' genes that were significantly up- or down-regulated by drought stress in cassava, including the ABA receptor MePYL2.
-
APETALA 2/ethylene-responsive element binding factor (AP2/ERFs) is a broad set of plant-specific TFs composed of four primary subfamilies: AP2, RAV, ERF, and dehydration-responsive element-binding protein (DREBs)[106]. The AP2/EREBP stimulates the expression of abiotic stress-responsive genes by specifically binding to the DRE/CRT cis-acting element (A/GCCGAC) in their promoter regions[107]. They participate in a variety of biological processes, including growth, development, hormone and stress responses. Increased AP2/ERF expression, particularly in the DREB, ERF, and RAV subfamilies, improves drought stress tolerance, making them suitable or possible candidate genes for crop improvement or genetic engineering[108, 109]. Up-regulation of AP2/ERF TFs, for example, improved drought tolerance in rice, wheat, transgenic tobacco, and Arabidopsis by increasing photosynthesis, ABA accumulation, proline biosynthesis, and ROS scavenging[109−111]. ABI3/VP1 (RAV) is another AP2/ERF-related TF that positively modulates drought tolerance in plants via ABA pathways[112]. Drought tolerance in rice[113] and wheat[114] was improved by up regulation of DREBs TFs. DREB2A and DREB2B in Arabidopsis operate as transcriptional activators in the ABA-independent (ABA-I) pathway via RD29A[ 19, 37,115]. DREB1A isolated from Arabidopsis improved drought resistance in transgenic rice[27].
The roles of these AP2/ERF TFs in cassava drought stress response have also been documented. For example, Liao et al.[116] demonstrated that multiple AP2/ERF subfamilies play important roles in the control of ethylene- and water-deficit stress-induced leaf abscission in cassava. These included MeERF1, MeERF4, MeCRF10, MeEBE, MeESE3, MeEDF1 (RAV TF), MeRAP2.4, MeERF12, MeRAP2.6, MeCRF9, MeERF9, and MeCRF11[116]. Similarly, Fan et al.[117] discovered potential MeERFs genes that were up regulated by drought stress in cassava leaves and roots. MeERF46, MeERF56, MeERF75, MeERF35, MeERF98, MeERF133, and MeERF136 were found to be up regulated in roots, while MeERF70, MeERF17, MeERF40, MeERF116, MeERF100, and MeERF128 were found to be up regulated in leaves[117]. Ren et al.[118] related higher expression of ethylene signaling-related gene families such as MeERF6, MeERF10, MeERF11, MeERF13, MeEIL1, MeERS1 and MeERS2 with enhanced accumulation of trehalose and proline contents in cassava leaves, stems, and roots under drought stress treatment. Trehalose and proline act as compatible solutes and perform various activities in plant cells to protect them from abiotic stressors[119]. MeERF1 was similarly strongly induced in cassava by drought stress treatment[17]. Yan et al.[60] demonstrated the involvement of one cassava RAV TF candidate gene (MeRAV5) in cassava drought tolerance via H2O2 regulation and enhanced lignin buildup. MeRAV5 increased the activities of peroxidase (MePOD) and cinnamyl alcohol dehydrogenase (MeCAD15), both of which alter H2O2 and accumulation of endogenous lignin, which are significant in drought stress tolerance in cassava.
Fu et al.[19] observed no differences in DREB2A/B gene expression levels in cassava during PEG-induced dehydration stress. However, Orek et al.[4] recorded differences in the expression of DREB1A, DREB2A/B, and RD29A/B genes in drought-tolerant and susceptible cassava genotypes exposed to different levels of water deficit treatments. Fu et al.[19] also analyzed another DREB2 member, DREB2C, which is linked in an ABA-insensitive pathway and has been shown to be co-expressed with a NAC protein, RD19, in response to dehydration and but not triggered by ABA in cassava. Under PEG-induced dehydration stress, MeRAP2.11 and MeRAP2.4 (another AP2/ERF family) for ethylene were also over expressed in cassava roots[19]. Other AP2/EREBP family members, notably MeSHN1, MeRAP2.4, MeANT, and MeABR1, have also been reported to be associated with drought stress response in cassava via hormones such as ethylene but not ABA[105].
-
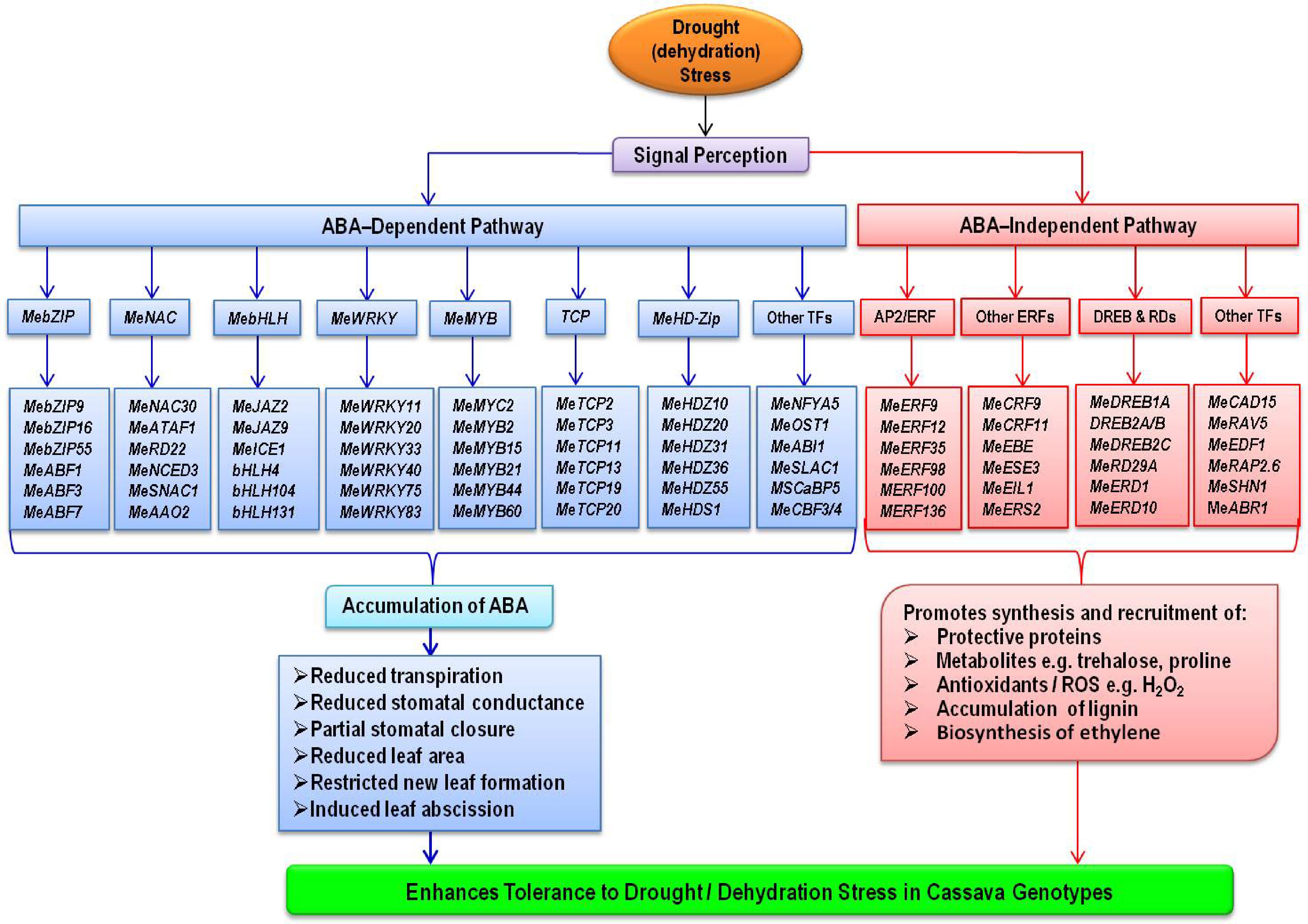
Expression of cysteine proteinase RD19 is not induced by ABA, but the gene is sensitive to dehydration stress[120]. Lokko et al.[43] identified RD19 as one of the unique expressed sequence tags (ESTs) that encode proteins with known involvement in cassava drought responses. Under PEG-induced dehydration stress, up regulation of RD19 was also observed in cassava[19]. Drought, natural and dark-induced senescence, and ABA all increase the expression of ERD1, which encodes a Clp protease regulatory subunit[120]. One of the unique ESTs that encode proteins with known involvement in drought responses in cassava was Precursor to Early Response to Desiccation 1, ERD1[43]. Delta1-pyrroline-5-carboxylate synthase (P5CS), a rate-limiting enzyme in proline biosynthesis, showed consistent expression patterns with MeERD1 in cassava under drought stress, implying that MeP5CS may participate in an ABA-independent pathway in cassava via ERD1[19]. Figure 1 depicts an overview of various ABA-dependent and ABA-independent transcription factors and related genes involved in drought tolerance in cassava as reviewed above.
-
Under water deficit conditions, ABA reduces transpirational water loss from cassava leaves, promotes partial stomatal closure, reduces leaf area by restricting new leaf formation or expansion and induces leaf abscission. These morpho-physiological responses are driven by a cascade of genes or transcription factors involved in ABA signaling pathways. This review article identified candidate transcription factors within ABA pathways that could be exploited to introgress drought tolerance traits not only in susceptible cassava genotypes but also other crop species. These included genes in AREBs/ABFs, NACs, bHLH, WRKY, MYC/MYB, HD-Zip, TCP, HSFs and AP2/ERFBPs families as well as NFYA5, SLAC1, ABI1, SCaBP5, PKS3, PYR1, GRXs, AP2/ERFs, DREB1A, DREB2A/B, RD29A/B, RD19 and ERD1. They can be considered for development of molecular markers for marker-assisted selection and also candidates for genetic engineering for drought tolerance.
-
The author thanks Dr. Evans Nyaboga, department of biochemistry at the University of Nairobi for his constructive feedback.
-
The author declares that there is no conflict of interest.
-
Received 6 June 2023; Accepted 9 August 2023; Published online 4 September 2023
- Copyright: © 2023 by the author(s). Published by Maximum Academic Press on behalf of Hainan University. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Orek C. 2023. A review of the functions of transcription factors and related genes involved in cassava (Manihot Esculenta Crantz) response to drought stress. Tropical Plants 2:14 doi: 10.48130/TP-2023-0014
A review of the functions of transcription factors and related genes involved in cassava (Manihot Esculenta Crantz) response to drought stress
- Received: 06 June 2023
- Accepted: 09 August 2023
- Published online: 04 September 2023
Abstract: Cassava navigates drought stress via diverse mechanisms including avoidance, tolerance, resistance or recovery from effects of drought. The crop's inherent tolerance to drought stress is underpinned by a set of genes involved in several molecular pathways. Among these include transcription factors (TFs) with key roles in abscisic acid (ABA) signaling pathways. ABA is a ubiquitous phytohormone that is critical in plant growth and development processes as well as responses to abiotic stresses such as drought. This review focuses on and summarizes the current developments in the identification, characterization and functions of TFs and related genes (RGEs) implicated in ABA pathways that regulate cassava's response to drought stress. The different drought-induced experiments set up either in the field or controlled environments and omics approaches applied by researchers for gene discovery and characterization are highlighted. The roles of these drought-induced genes in other crops or plants are compared with cassava. The review reveals functions of key candidate TFs and REGs including AREBs/ABFs, NACs, bHLH, WRKY, MYC/MYB, HD-Zip, TCP, HSFs, AP2/ERFBPs, NFYA5, SLAC1, ABI1, SCaBP5, PKS3, PYR1, AP2/ERFs, DREB1A, DREB2A/B, RD29A/B, RD19, ERD1 among others. These genes are potential molecular markers that could aid in rapid introgression of drought tolerance traits not only in farmer-preferred and drought susceptible cassava genotypes, but also in other crops for improved production. Through this omics-based drought-mitigation, the negative effects of climate change could be reduced.
-
Key words:
- Cassava /
- Climate change /
- Drought stress /
- ABA /
- Molecular markers