-
Plants exhibit diverse natural associations with microorganisms through various mechanisms. These associations with spatially and temporally complex land plants could be in the form of pathogens, PGPR (plant growth-promoting rhizobacteria) and endophytes[1]. The symbiotic relationship between endophytic bacteria and plants plays a crucial role in enhancing seed germination, fostering plant growth, boosting crop yield, influencing secondary metabolite production, improving disease resistance, and bolstering tolerance to environmental stressors like drought and salinity[2]. Given their widespread presence in plants and their ecological and economic significance, it is imperative to elucidate the intricate interactions among bacterial endophytes, their host plants, and agricultural practices. In recent decades, investigations into endophytic bacteria have predominantly concentrated on delineating their advantageous effects and ecological dynamics, aiming to enhance comprehension regarding the intricate interactions between bacterial endophytes and their respective host plants[3, 4].
Soil salinization exerts a substantial detrimental effect on agricultural productivity worldwide, affecting both irrigated and non-irrigated regions. It induces osmotic stress, water scarcity, stomatal closure, and diminished leaf growth. Additionally, salinity in the soil leads to the depletion of vital nutrients like potassium (K+). Elevated Na+ inside plants can decrease plant photosynthetic rates and biomass accumulation[5−7]. Most crop and forage plants that feed the global population are sensitive to high salt concentration in soils[8]. Investigating the community structure of endophytic bacteria in saline environments could reveal the distribution characteristics of microbial populations and enable evaluations of homeostasis in the tissues of host plants. Advanced sequencing technologies have provided extensive data sources, which have facilitated the study of microbial species, community structure, community function, and genetic diversity. Over the past ten years, significant progress has been made in studies on environmental microbial diversity based on these technologies.
This work aimed to isolate bacterial endophytes from a halotolerant plant, Chloris virgata, a member of the grass family Poaceae. We examined potential in vitro beneficial physiological traits induced by the endophytes in plants under normal or salt-stressed conditions. In particular, the following traits of bacterial endophytes were analyzed in this study: siderophore production, phosphate solubilization, synthesis of auxin, capacity to promote plant growth, resistance to antibiotics, ACC deaminase synthesis, and the ability to increase plant tolerance to salinity stress.
-
Healthy plant samples of C. virgata were randomly collected during the month of April−May 2018 from five sites around the agricultural fields situated at the Institute of Agriculture Sciences, Banaras Hindu University, Varanasi, India.
Isolation of endophytic bacteria and biochemical characterization
-
The endophytic bacteria were isolated from the roots, rhizomes, and leaves of C.virgata plants following the surface sterilization protocol[9]. The plant parts were thoroughly washed with running tap water to remove soil particles and attached microbes. Root, rhizome and leaves were individually surface sterilized by serial washing in 70% ethanol (1 min), 2% sodium hypochlorite solution (available Cl−) for 3−4 min and 70% ethanol (60 s) followed by rinsing three times with sterile distilled water. Isolation was performed using the direct plating method on nutrient agar (NA) medium supplemented with 2% sodium chloride (NaCl). The sterilized leaves were cut into 0.5 cm × 0.5 cm sections while root and rhizome were cut into 2 mm sections. A minimum of five sections were placed on NA plates and incubated at 35 ± 2 °C for 24−72 h. The plates were checked every 24 h for bacterial colony emergence. The bacterial colonies were selected, sub-cultured, purified, and used for further studies. The bacterial isolates were characterized after 48 h for the following traits: colony color, form, elevation, margin, surface, opacity, and texture. Biochemical tests such as indole production, methyl red and Vogues-Proskauer tests, citrate utilization, catalase and oxidase production tests, nitrate reduction, and oxidative fermentation tests were performed for the endophytic isolates.
Plant growth promotion traits
-
Pure cultures of the bacterial isolates were inoculated onto solid diagnostic media supplemented with respective substrates to test enzyme activity. Protease, cellulase, lipase and amylase activity were assessed on Gelatin Yeast extract Peptone agar (GYP agar), Carboxymethyl cellulose agar (CMC agar), Tween 20 agar, and starch agar, respectively. The agar plates for enzyme activity testing were spot inoculated with the bacterial isolates and incubated at 30 ± 2 °C for 24−36 h. Proteolytic activity was detected by flooding the plates with saturated ammonium sulfate solution. The formation of a clear zone around bacterial colonies indicated substrate breakdown by proteases. Cellulase activity was evaluated by adding 0.2% (w/v) Congo red solution to the plates followed by washing with 5 M NaCl solution. The appearance of clear zones around colonies against a red background signified cellulase activity. Amylolytic activity was assessed by flooding the plates with 2% iodine solution and observing for clear zones, indicating substrate hydrolysis. Lipase activity was denoted by crystallization around bacterial colonies[10−12].
Indoleacetic and Gibberellic Acid production
-
Nutrient broth (NB) media supplemented with 0.01% w/v tryptophan was used for IAA production and analyzed with Salkowski reagent (35% HClO4 50 mL, 0.5 M FeCl3 1 mL). Uninoculated growth medium was used as the negative control. The IAA concentration in the culture was estimated based on the IAA standard curve[13]. Gibberellic acid was estimated following the modified protocol of Graham & Thomas[14].
Phosphate solubilization
-
Isolates were spot inoculated onto the National Botanical Research Institute's phosphate growth medium (NBRIP) containing 2% w/v tricalcium phosphate [Ca3(PO4)2] and incubated at 30 °C for 7−10 d. Formation of clear zones around growth indicated the phosphate solubilizing activity of bacterial isolates[15].
ACC deaminase activity
-
Endophyte bacterial strains were cultured in nutrient broth to test the activity of ACC deaminase. Bacterial cells were harvested (centrifuged at 5,000 rpm for 15 min), washed with sterilized 0.1 M Tris-HCl (pH 7.5) and were inoculated onto a medium of minimal saline agar. A medium with the addition of 0.2% NH4(SO4)2 was used for positive control. DF (Dworkin and Foster) medium was used as a negative control. The cultures were incubated for 36 h at 28 ± 2 °C. The appearance of endophytic bacterial colonies in DF minimal saline medium was evaluated as a positive control[16].
Siderophore, ammonia, and HCN production
-
The Chrome azurol assay (CAS) was followed for siderophore estimation[17]. All bacterial isolates underwent spot inoculation onto CAS agar plates and were then incubated at 30 °C for 24 h. The observation of a yellow to light orange halo formation around the colonies served as an indicator for siderophore production. Siderophore production was also assessed by using a modified microtitre plate method[18]. The bacterial cultures were cultivated in broth, and subsequently, 100 µL of the supernatant from each bacterial culture was introduced into individual wells of a microplate, followed by the addition of 100 µL of CAS reagent. Following the designated incubation period, absorbance readings were measured at 630 nm using a microplate reader (Agilent BioTek Synergy HTX Multimode Reader).
$\begin{aligned}&{\rm PSU\;({\mathrm{\%}}\;siderophore\;unit)}=\\&\quad\left[ \dfrac{Absorbance\;(re f erence)-Absorbance\;(sample)}{Absorbance\;(re f erence)}\right]\,\times\, 100{\mathrm{\%}}\end{aligned} $ To assess ammonia production, 1 mL of Nessler's reagent was introduced to bacterial cultures cultivated in peptone water. The emergence of a light yellow to brown coloration indicated the presence of ammonia[19]. For the examination of hydrogen cyanide (HCN) production, the bacterial cultures were inoculated onto NB medium supplemented with 0.4% glycine. Additionally, Whatman No.1 filter paper strips soaked in 0.5% picric acid in 2% sodium carbonate were placed within the Petri dish lid, which was then sealed with Parafilm. Incubation at 28 °C for duration of four days was ensued. A change in the color of the strips from yellow to brown signified a positive result for HCN production[20].
Nitrogen free growth
-
Bacteria cultured in Jensen's medium (Sucrose 20 g; K2HPO4 1.0 g; MgSO4·7H2O 0.5 g; NaCl 0.5 g; FeSO4·7H2O 0.1 g; Na2MoO4 0.005 g; CaCO3 2.0 g; Agar 15.0 g) were incubated for 7 d at 28 ± 2 °C. The emergence of endophytic bacterial colonies in Petri dishes during the incubation period indicates that the bacterial cultures possess the ability to grow in N-free medium[21].
In vitro evaluation of stress response to osmotic and salinity
-
To characterize salt tolerance, the isolates were cultivated at 35 ± 2 °C for 72 h in NA medium with varying NaCl concentrations (5%, 10%, 15%, and 20%). The assessment of osmotic stress tolerance involved the isolates' growth observation in nutrient agar medium supplemented with polyethylene glycol (PEG-6000) at 35 ± 2 °C for 24 h. The osmotic potentials in the media ranged from −0.15 MPa to −1.03 MPa[22].
Molecular identification of endophytic isolates
-
A total of nine out of 13 isolates were selected for identification based on their PGP activities. Genomic DNA was extracted according to Wilson[23]. The isolated genomic strains were analyzed on 0.8% agarose gel (Himedia) and amplification of the 16S rRNA (~1,500 bp) gene was carried in PCR using universal primers [forward primer (27F) AGAGTTTGATCCTGGCTCAG, Reverse primer (1492R), CGGTTACCTTGTTACGACTT (Eurofin Genomics, India)]. PCR was performed with 25 µL reaction mixture composed of PCR buffer 2.5 µL; MgCl2 1.5 µL; 0.2 mm dNTPs 2 µL; Primers each 1.25 µL; Taq DNA polymerase 0.25 µL (Thermo Fischer Scientific); template DNA 2 µL; PCR water 14.75 µL. The PCR conditions were as follows: initial denaturation 94 °C for 4 min, followed by 30 cycles of 30 s denaturation at 94 °C, 1 min annealing at 63 °C, 1 min extension at 72 °C and 7 min final extension at 72 °C. The PCR products were purified before they were sent for sequencing.
Phylogenetic characterization
-
The PCR products of the endophytes were sequenced by BIOKART INDIA Pvt. Ltd (Bengaluru, India) by Sanger's dideoxy nucleotide sequencing method. All obtained sequences were compared with those in the GenBank database by using the BLASTN search program. Similar sequences were further aligned by CLUSTALW. Phylogenetic analysis was conducted by the neighbor-joining method. Bootstrap analysis was performed with 1,000 replications to determine the support for each clad with grouping by the neighbor-joining method. A phylogenetic tree was constructed based on evolutionary distance data by using MEGA 11[24].
Nucleotide accession number
-
The 16S rRNA nucleotide sequences acquired have been submitted to the National Center for Biotechnology Information (NCBI) GenBank to secure accession numbers. The unique accession numbers assigned to the bacterial isolates are MW680956 through MW680964.
Statistical analyses
-
All samples underwent triplicate analysis, and the data were reported as the mean value accompanied by the standard deviation for each data point. Data processing and visualization software options include GraphPad Prism (version 10.1.0), Molecular Evolutionary Genetics Analysis (MEGA version 11.0.13), and InteractiVenn for Venn diagram analysis[25].
-
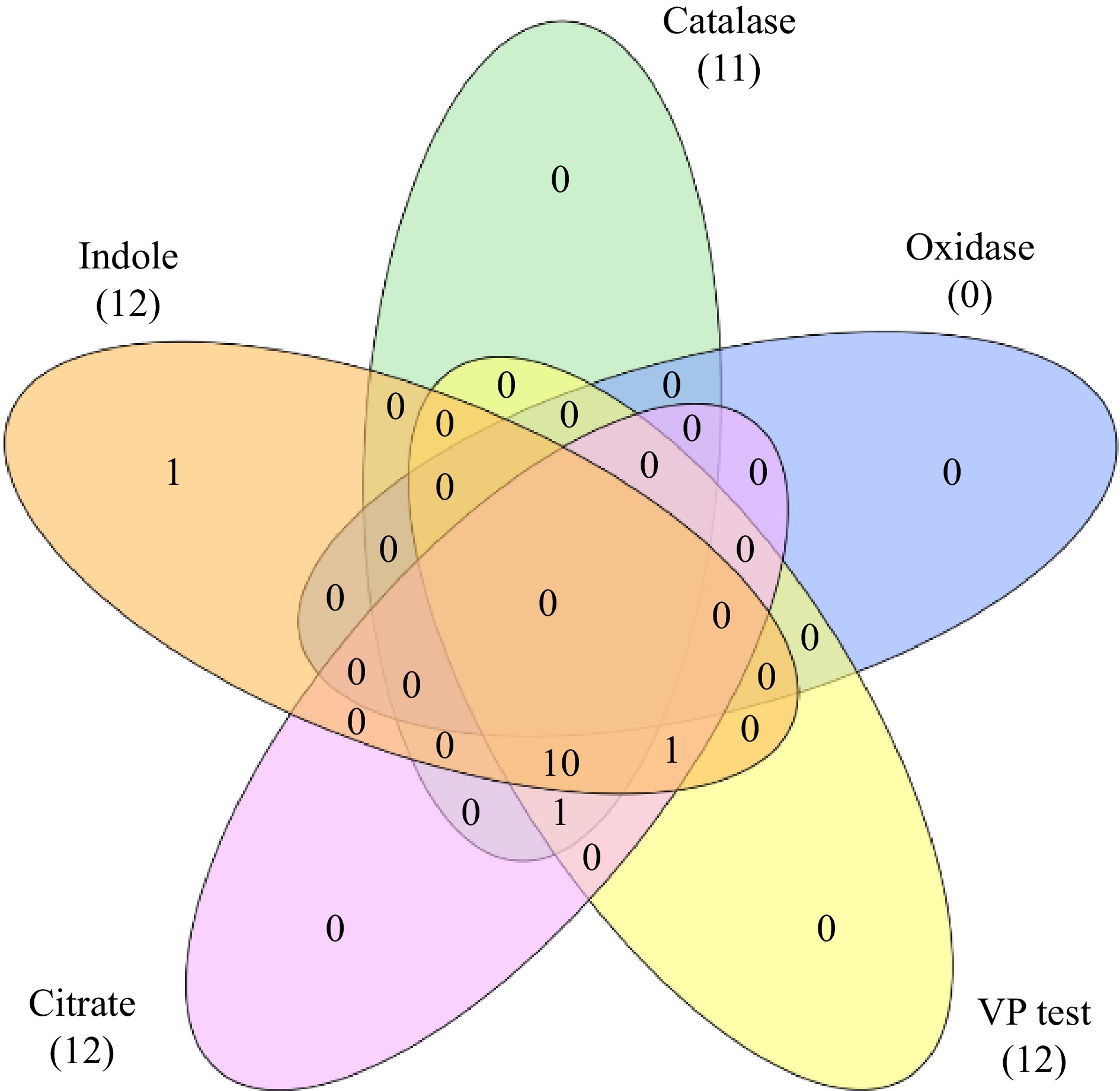
A total of 43 endophytic bacterial isolates were recovered from different parts of C. virgata. The number of bacterial isolates obtained from leaf, rhizome, and root were 9, 12, and 22 respectively. However they were grouped as per their morphological similarity (appearance of colonies) for further study into 13 strains (Table 1). All the isolates were oxidase negative but catalase positive and most of the isolates exhibited positive result for Voges-Proskauer test, oxidative fermentation, citrate utilization and nitrate reduction test (Table 2; Fig 1). The leaf isolate was indole negative, whereas all the root and rhizome isolates were indole positive.
Table 1. Morphological characteristic of endophytic bacterial isolates from Chloris virgata.
Isolates Colour Surface Shape Edge/margin Elevation Opacity CHVLB1 Off-white Shiny Circular Smooth Raised Opaque CHVRB1 Off-white Irregular − Lobate Flat Opaque CHVRB2 Off-white to pink Irregular − Undulate Raised Opaque CHVRB3 Off-white to pink Shiny Circular Lobate Raised Opaque CHVRB4 Off-white to pink Mucoidal, glistening − Entire Raised Opaque CHVRB5 Off-white − − Lobate Flat Opaque CHVRTB1 Off-white Dull Circular Entire Raised Opaque CHVRTB2 Yellow − Irregular Undulate Raised Opaque CHVRTB3 Off-white − Irregular Undulate Raised Opaque CHVRTB4 Off-white − − − Flat Opaque CHVRTB5 Off-white Irregular − Lobate Flat Opaque CHVRTB6 Off-white Irregular − Lobate Raised Opaque CHVRTB7 Off-white Slimy, glistening Circular Entire Raised Opaque * CHVLB: Chloris virgata Leaf bacteria; CHVRB: Chloris virgata Rhizome bacteria; CHVRTB: Chloris virgata Root bacteria. Table 2. Biochemical characterization of endophytic bacteria.
Isolates Indole production Catalase Oxidase Methyl Red test Voges-Proskauer test Citrate utillisation Oxidative fermentation NO3− With oil Without oil CHVLB1 − + − − + + + + + CHVRB1 + + − − + + + + − CHVRB2 + + − − + + + + + CHVRB3 + + − − + + − + + CHVRB4 + + − − + + + + + CHVRB5 + + − − + + + + + CHVRTB1 + + − − + + + + + CHVRTB2 + + − − + + + + − CHVRTB3 + + − − + + + + + CHVRTB4 + + − + + + + + + CHVRTB5 + − − − + + + + − CHVRTB6 + − − + − − + + + CHVRTB7 + + − − + + + + + (+) = presence of activity; (−) = absence of activity. Plant growth promoting traits
-
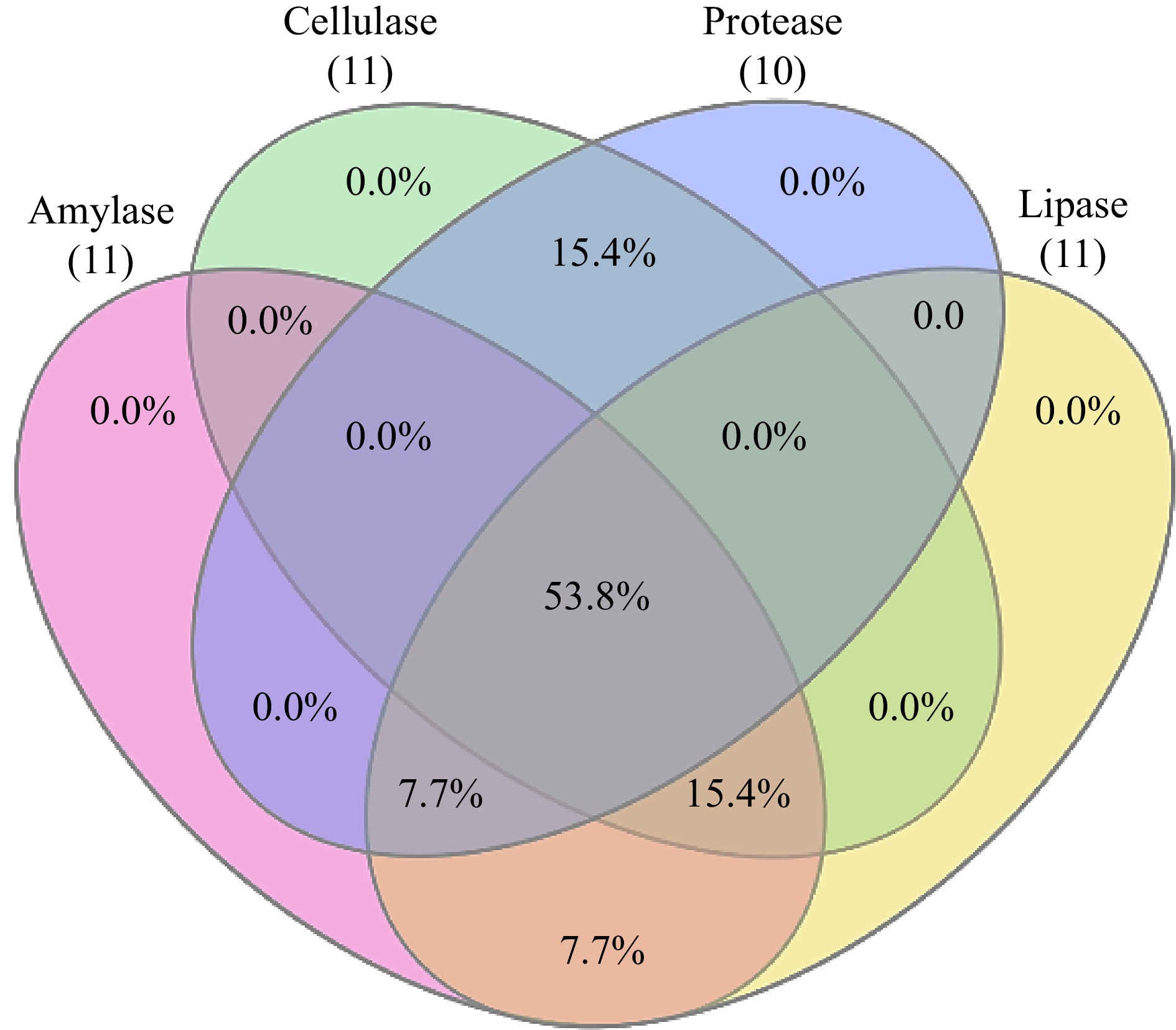
Amylase activity was present in 84.6% of the isolates. All root isolates except CHVRTB3 and CHVRTB7 (84.6%) isolates were positive for cellulase activity, except isolates CHVRTB4 and CHVRTB7 all isolates had shown cellulase activity. Protease activity was evident for (69.2%) isolates CHVLB1, CHRB2–CHVRB5; CHVRTB1, 2, 3 and 7. All isolates except CHVLB1 and CHVRTB3 were positive for lipase activity (Table 3, Fig 2).
Table 3. Qualitative observation of PGP traits of the endophytic isolates.
Isolates Enzyme activity Phosphate utillisation Siderophore production Ammonia production HCN
productionAcc deaminase Nitrogen free growth Amylase Cellulase Protease Lipase TCP DCP CHVLB1 − +++ +++ − − + +++ ++ − + +++ CHVRB1 +++ ++ − +++ − + + +++ − + + CHVRB2 +++ +++ ++ +++ + + ++ +++ − + + CHVRB3 +++ +++ +++ +++ + + + +++ − + + CHVRB4 +++ +++ +++ +++ + + +++ +++ − + + CHVRB5 +++ +++ ++ +++ − + − +++ − + + CHVRTB1 +++ +++ +++ +++ − + +++ ++ − + +++ CHVRTB2 +++ +++ +++ +++ + + +++ ++ − + +++ CHVRTB3 − +++ +++ − + + +++ ++ − + +++ CHVRTB4 +++ − − +++ + − +++ ++ − − − CHVRTB5 +++ + − +++ − + − +++ − − + CHVRTB6 +++ + − +++ − − + +++ − − + CHVRTB7 + − +++ + + − − +++ − + + * HCN: Hydrogen cyanide; TCP: Tricalcium phosphate, DCP: Dicalcium phosphate; Number of (+) sign indicates strength of activity, (−) indicates absence of activity. IAA production of all isolates ranged between 0.033 to 0.054 μg·mL−1 (without tryptophan supplement) and between 11.53 to 29.53 μg·mL−1 (with tryptophan supplement). The isolated strains demonstrated the ability to biosynthesize indole-3-acetic acid (IAA) analogs in a minimal liquid medium containing 100 µg·mL−1of tryptophan (Table 4). All strains exhibited the ability to produce > 10 µg·mL−1 of IAA. Specifically, 38.46% of the isolates produced over 20 µg·mL−1of IAA, while 61.53% of the isolates yielded > 20 µg·mL−1of IAA analogs. Notably, the root endophyte CHVRTB4 (identified as Bacillus sp.) displayed the highest average IAA production at 29.53 µg·mL−1, whereas CHVRTB3 (again Bacillus sp.) exhibited the lowest average production at 11.53 µg·mL−1.
Table 4. Quantitative observation of PGP traits of the endophytic isolates.
Isolates Indole acetic acid production (µg·mL−1) Gibberellic acid production (µg·mL−1)
(540 nm)Siderophore production (CAS ASSAY)
% PSUAmmonia production
(µg·mL−1)Without trp With trp CHVLB1 0.033 ± 0.006 13.50 ± 0.004 22.22 ± 0.003 32.71 ± 0.008 35.45 ± 0.004 CHVRB1 0.036 ± 0.006 16.91 ± 0.008 146.72 ± 0.004 41.05 ± 0.012 71.28 ± 0.031 CHVRB2 0.039 ± 0.003 17.62 ± 0.006 70.64 ± 0.009 49.40 ± 0.003 95.27 ± 0.009 CHVRB3 0.038 ± 0.005 20.76 ± 0.003 74.92 ± 0.007 49.30 ± 0.003 98.33 ± 0.006 CHVRB4 0.044 ± 0.004 17.84 ± 0.004 73.93 ± 0.009 48.96 ± 0.006 94.86 ± 0.005 CHVRB5 0.028 ± 0.009 23.95 ± 0.005 97.64 ± 0.008 8.16 ± 0.003 73.64 ± 0.032 CHVRTB1 0.038 ± 0.004 12.19 ± 0.007 *c149.03 ± 0.012 49.08 ± 0.002 69.05 ± 0.003 CHVRTB2 0.035 ± 0.014 18.14 ± 0.004 136.84 ± 0.006 27.50 ± 0.025 97.29 ± 0.031 CHVRTB3 0.037 ± 0.01 11.53 ± 0.002 103.57 ± 0.005 48.83 ± 0.002 74.89 ± 0.015 CHVRTB4 0.032 ± 0.007 *b29.53 ± 0.008 96.33 ± 0.005 *d69.77 ± 0.063 *e276.13 ± 0.13 CHVRTB5 *a0.054 ± 0.012 16.42 ± 0.008 92.05 ± 0.003 36.55 ± 0.10 79.97 ± 0.01 CHVRTB6 0.033 ± 0.016 22.96 ± 0.004 102.26 ± 0.005 32.04 ± 0.004 70.09 ± 0.007 CHVRTB7 0.037 ± 0.02 24.25 ± 0.005 57.79 ± 0.003 11.01 ± 0.002 66.54 ± 0.003 *a = highest IAA produced without trp supplement; *b = highest IAA produced with trp supplement; *c = highest GA3 concentration produced; *d = highest PSU released; *e = highest concentration of ammonia released. Similar to IAA production all isolates tested produced GA3. Gibberellic acid production of all isolates ranged between 22.2−149.03 µg·mL−1 with 92.30% of the isolates producing > 50.0 µg·mL−1, highest mean being 149.03 µg·mL−1 (CHVRTB1 identified as Glutamicibacter sp.), CHVRB1 (146.72 μg·mL−1) as the second highest and lowest mean being 22.22 µg·mL−1 by CHVLB1. All isolates had shown positive result for qualitative ammonia production test (Fig. 3). The quantitative assessment revealed ammonia production levels ranging from 35.45−276.13 µg·mL−1. Specifically, 53.84% of the isolates exhibited ammonia production within the range of 70−90 µg·mL−1, while 38.46% of the isolates demonstrated production levels between 90−270 µg·mL−1. Notably, isolates CHVRTB1 and CHVLB1 displayed the highest (276.13 µg·mL−1) and the lowest (35.45 µg·mL−1) concentrations of ammonia production, respectively.
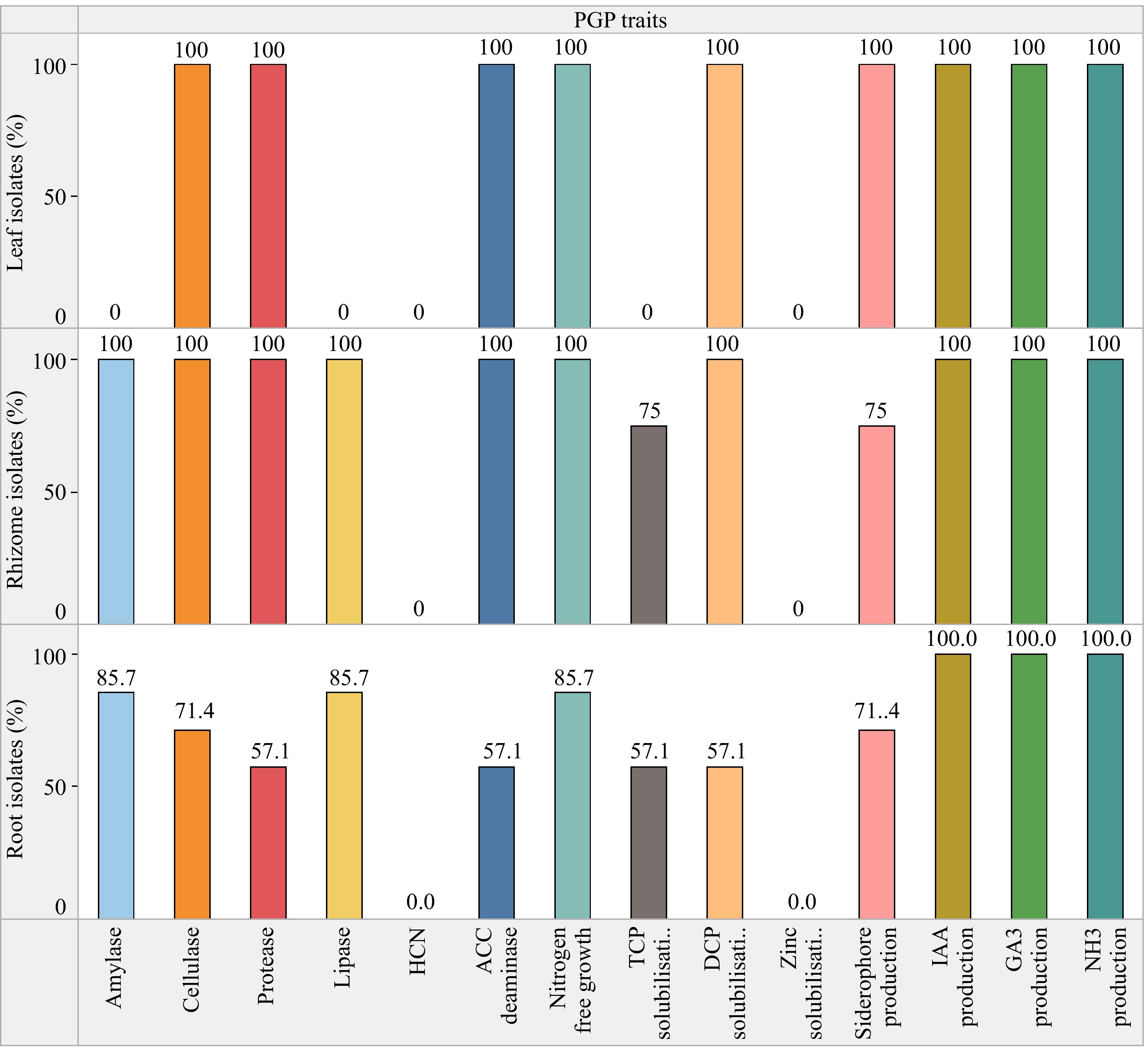
Conversely, a lower percentage of the examined bacterial endophytic isolates, specifically 76.9% and 61.5%, exhibited positive outcomes for siderophore production and phosphate solubilization respectively. Phosphate solubilization was performed using both tricalcium (TCP) and dicalcium (DCP) phosphates as substrate. A total of 76.9% isolates showed DCP solubilization while 53.8% solubilized TCP. Zinc solubilization was altogether absent in all the isolates. Siderophores function by chelating iron and other metals, facilitating their absorption by plants. Additionally, they play a role in disease management by outcompeting phytopathogens for essential trace metals. Percent siderophore unit (PSU) was calculated for the isolates which was maximum for isolate CHVRTB4 (69.77%) and least for CHVRB5 (8.16%), unexpectedly leaf isolate CHVLB1 showed 32.71% siderophore units (Fig. 4). No bacterial isolates tested exhibited hydrogen cyanide (HCN) production. The ACC deaminase activity, known for aiding plants in resilience against both biotic and abiotic stress, was identified in 10 out of the 13 bacterial isolates. The majority of the isolated endophytic bacteria possessed more than three PGP traits out of the eight tested traits. The isolate CHVRTB4 obtained from the root exhibited majority of the PGP traits. When compared to the isolates from roots and leaves, the rhizome isolates showed positive outcome for the majority of the plant growth-promoting traits tested.
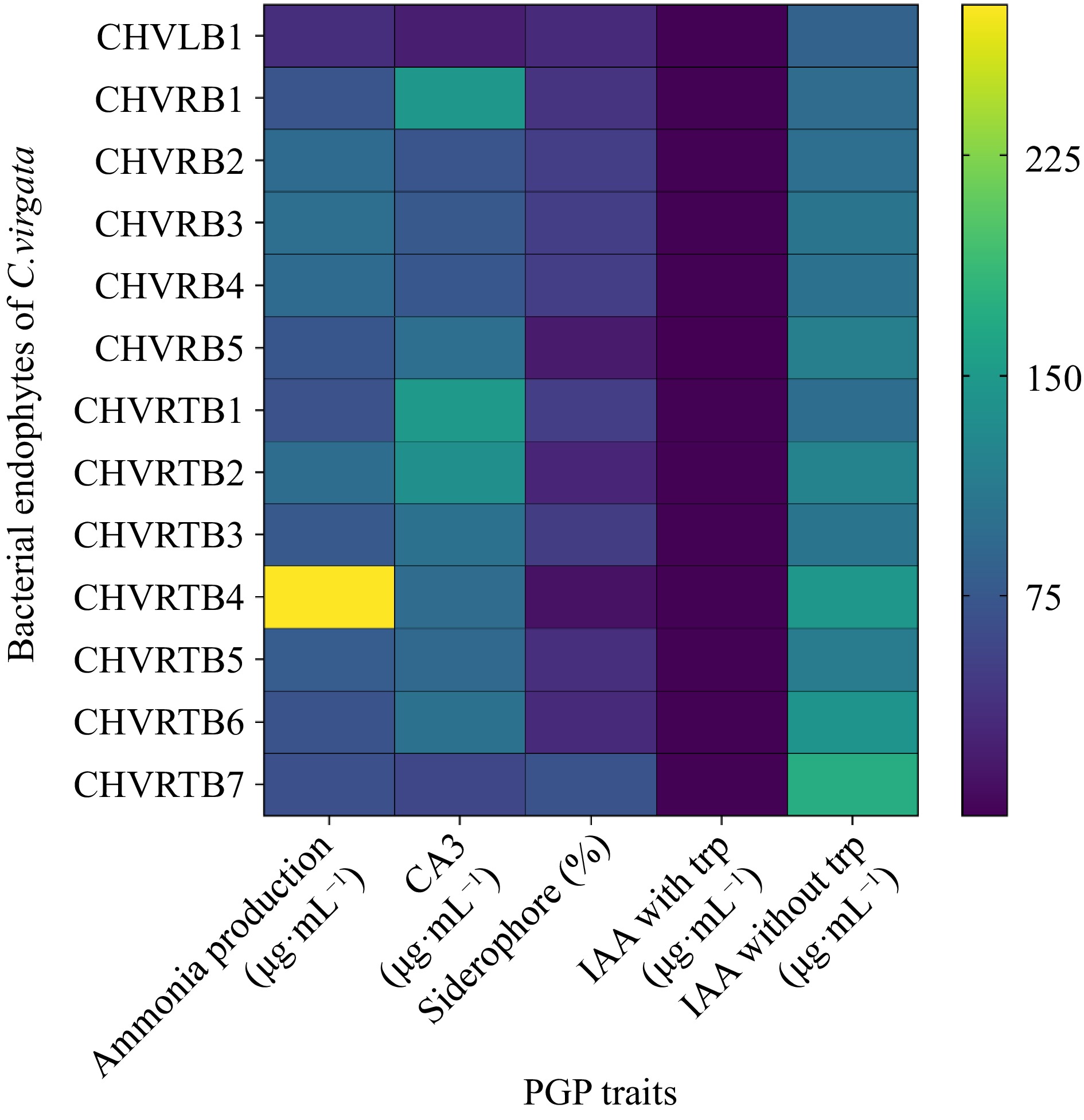
Figure 4.
Heatmap for plant growth promoting traits of the bacterial endophytes was prepared in terms of color intensity by normalisation of mean values of the results.
In vitro evaluation of bacterial response against salinity and osmotic stress
-
All 13 isolates exhibited the capacity to tolerate and thrive in NA medium supplemented with 5% NaCl. However, among these isolates, CHVRB1, CHVRTB4, and CHVRTB6 demonstrated limited growth in NA medium supplemented with NaCl, only up to 10%. No observable growth occurred at 15% and 20% salt concentrations. Regarding osmotic stress, when utilizing PEG 6000, 12 out of the 13 isolates demonstrated growth up to a water potential of –1.02 MPa (Table 5). In the context of bacterial osmotic stress tolerance, research has elucidated the biological mechanisms underlying the adaptation and survival of microorganisms in extreme environments. Under osmotic stress conditions, bacterial cells accumulate small compatible solutes, known as osmolytes. These include amino acids such as glutamate, glutamine, proline, alanine, quaternary amines like glycine betaine, and sugars like sucrose, trehalose, and polyglucosyl granules. These osmolytes play a crucial role in improving cell growth under adverse osmotic conditions, serving as effective osmoprotectants.
Table 5. Screening for salinity and osmotic stress tolerance on minimal DF (Dworkin and Foster) media.
Isolates 5%
NaCl10%
NaCl15%
NaCl20%
NaCl0.15
MPA0.49
MPA1.03
MPACHLB1 + + − − + + + CHRB1 + + − − + + + CHRB2 + + − − + + + CHRB3 + + − − + + + CHRB4 + + − − + + + CHRB5 + − − − + + +++ CHRTB1 + + − − + + + CHRTB2 + + − − + + +++ CHRTB3 + + − − + + ++ CHRTB4 + + − − + + +++ CHRTB5 + + − − + + ++ CHRTB6 + − − − + + ++ CHRTB7 + − − − + + +++ Number of (+) sign indicates the increased appearance of colonies on media, (−) indicates absence of activity. Phylogenetic analysis of Chloris virgata bacterial endophytes based on 16s rRNA sequencing
-
All isolates exhibited 96%–100% similarity to sequences in the National Center for Biotechnology Information (NCBI) database. The phylogenetic analysis was divided into two groups according to genus: Bacillus and Glutamibacter.
Eight bacteria belonged to the Bacillus group (isolates CHVRB1, CHVRB2, CHVRB3, CHVRB4, CHVLB1, CHVRTB2, CHVRTB3, and CHVRTB4) and one belonged to the Glutamicibacter group (isolate CHVRTB1). The analysis is represented by order Bacillales and Micrococcales. The 16s rDNA sequences were deposited in the GenBank with accession numbers MW680956–MW680964 (Table 6). The resulting phylogenetic tree of bacterial isolates revealed that isolates and reference sequences were clustered according to established taxonomic orders, with high bootstrap support (Fig. 5). The majority of the isolates were categorized under Bacillales order.
Table 6. Taxonomic identification and sequence homology of isolated bacterial endophytes with genus or species affiliation and corresponding identity percentage in the NCBI database.
Endophytic isolates Accession number Bacterial genus with highest similarity Accession number of closest match Identity % CHVLB1 MW680956 Bacillus sp. NR_116240.1 99.45 CHVRB1 MW680957 Bacillus sp. NR_148786.1 98.97 CHVRB2 MW680958 Bacillus sp. NR_112686.1 98.64 CHVRB3 MW680959 Bacillus sp NR_112686.1 99.75 CHVRB4 MW680960 Bacillus sp. NR_112686.1 99.84 CHVRTB1 MW680961 Glutamicibacter sp. NR_026190.1 99.21 CHVRTB2 MW680962 Bacillus sp. NR_113265.1 99.84 CHVRTB3 MW680963 Bacillus sp. NR_112686.1 99.92 CHVRTB4 MW680964 Bacillus sp. NR_157609.1 99.25 
Figure 5.
Neighbor-joining phylogenetic tree based on the partial sequence of the 16S rRNA gene of bacterial isolates from C. virgata and their related type strains. The evolutionary distances were computed using the p-distance. Bootstrap values are given at branch nodes and are based on 1,000 replicates.
-
In this study isolation of endophytes from C. virgata following surface sterilization resulted in 43 bacterial isolates where highest (37.2%) from root, rhizome (27.9%) and least from leaf (20.9%). However when grouped on morphological and biochemical characteristics and identified on molecular basis they fell only into two categorical orders (Bacillales and Micrococcales). Functional characterization of culturable endophytic bacteria is integral to enhancing our understanding of microbial communities existing in association with plants, such as endophytic bacteria. A significant proportion of the isolates exhibited notable enzymatic activities, including cellulase, amylase, lipase, and protease. These enzymes, encompassing cutinase, pectinase, cellulase, hemicellulase, protease, and lignin-peroxidases are predominantly linked to their involvement in penetrating plant tissues[26]. Phosphate-solubilizing bacteria, when utilized as inoculants, augment phosphorus absorption by plants, consequently enhancing crop yield. The capability to solubilize inorganic phosphate is frequently observed in rhizospheric bacteria. DCP solubilization was exhibited by 2/3 isolates, while 1/3 isolates could solubilize TCP. Siderophores sequester iron (Fe3+, Fe2+) from the environment, making it accessible for use by both microbial and plant cells. Moreover, siderophores possess the ability to chelate undesirable metal ions (like Zn, Cd, Cr, Al, and Pb) in the rhizosphere, thus alleviating potential toxicity for plants. Furthermore, siderophores play a pivotal role in plant defense by actively acquiring nutrients and outcompeting phytopathogens in a competitive manner[27]. Among the 13 bacterial endophytic isolates, 10 exhibited the ability to produce siderophores. The volatile secondary metabolite HCN is naturally synthesized by various soil microbes and endophytic bacteria[28]. Moreover, HCN possesses broad-spectrum toxicity against fungi, nematodes, insects, and plants due to the sensitivity of heme groups in the targeted eukaryotic cells[29]. However, none of the isolates displayed any detectable HCN production activity.
Plants benefit from ACC deaminase-producing PGPEs as they aid in alleviating various stress-related symptoms, stimulating growth, and supporting adaptation and survival. These beneficial microbes degrade ACC into ammonia and α-ketobutyrate, providing plants with nitrogen and the necessary energy for optimal growth under stressful conditions[30]. In this study bacterial endophytes were grown on DF minimal media to identify isolates able to synthesize ACC deaminase and grow in the medium. Positively all the isolates grew on DF medium. Jensen's N-free medium is usually used for isolation of diazotrophic microbes[31]. However here it was used to check the growth of the isolates in N-free medium thus attempting to establish their role as a free living diazotroph.
Phytohormone production raises tolerance of the plant to abiotic stresses and promotes plant growth[32]. Microbial IAA acts as a signaling molecule in plant-microorganism interactions. IAA production is known to be one of the most common traits among the endophytic bacteria. IAA has been associated with developmental processes of vascular tissue, adventitious root initiation, tropistic responses, apical dominance, and the development of flowers and fruits. It also influences functions like cellular differentiation, expansion, and division[33]. In this study the majority of the isolates were able to produce IAA, GA3, and ammonia. IAA production was exhibited by all the isolates. Gibberellic acid (GA) plays a pivotal role in plants for seed germination, elongation growth, flowering time regulation, and floral development. GA3 stands out as one of the key bioactive gibberellins, among others[34]. Bacillus spp. (B. siamensis, B. amyloliquefaciens, B. subtilis, B. licheniformis, etc.) have been emerging as the largest and most promising group of naturally occurring (PGPR and PGBEs) that enhance plant growth via GA[35]. GA3 production was comparatively higher in the root isolates.
A significant portion of plants can utilize only inorganic forms of nitrogen, such as ammonium and nitrate, and cannot directly utilize atmospheric nitrogen[36]. In this study the root and rhizome isolates of C. virgata were the primary ammonia producers.
Salinity stress is one of the major abiotic stresses directly responsible for drastic effects on the physical and chemical properties of soil but also suppresses the growth and assortment of soil microbiota, nematodes and crop plants, resulting in stunted plant growth that eventually lead to reduced crop production[37]. Remarkably, almost all endophytic bacteria (isolated and characterized in the present study) grew on nutrient agar medium supplemented with NaCl (5%−20%). All isolates tolerated upto 10% NaCl in the medium. The primary focus of the study centered on evaluating the plant growth-promoting (PGP) traits, as well as the tolerance of selected endophytes associated with C. virgata to salinity and osmotic stress. Out of the 13 isolates, nine were identified, with the majority (8) belonging to the Bacillales order, mainly Bacillus sp., and the remaining one to the Micrococcales order, identified as Glutamicibacter sp. A previous study on diversity of endophytic bacteria of C. virgata growing on oil contaminated sites showed Bacillus as dominant genera. The study in itself was a first on the community structure of endophytic bacteria in C. virgata. Bacillus sp. is the most common colonizing genus in the rhizosphere and roots of gramineous plants, which may be the cause of its predominance among hydrocarbon-degrading bacteria[38].
Traditionally, cultivation-dependent techniques were employed to analyze the community structure of endophytic microbes. Culture dependent methods are commonly cost-effective. However, a notable drawback of cultivation-dependent approaches in elucidating endophyte diversity lies in the bias towards fast-growing, ubiquitous species. This bias may result in the oversight of rare species with lower competitive strength and more specialized requirements, limiting the scope of functional analysis and the potential agricultural applications of these overlooked species[39, 40]. Conversely, when only a small amount of nucleic acid of suboptimal quality is obtainable from the source, the application of culture-independent molecular methods becomes challenging. The majority of bacteria are not amenable to cultivation, leading to a restricted understanding of the mechanisms governing the interactions between plants and beneficial endophytic bacteria. This limitation impedes the practical utilization of endophytes. Therefore, in the future employing a sophisticated and comprehensive omics approach to investigate both the structural and functional aspects of microbial communities can serve as a means to overcome this obstacle.
-
The findings reported here suggest that the endophytic bacteria isolated from C. virgata could have potential applications in plant disease management by promoting plant growth, mitigating stress, and enhancing disease resistance in host plants. It will also help in conducting detailed studies to understand the mechanisms behind the functional characteristics exhibited by the endophytic bacteria, such as enzyme activities, phosphate solubilization, siderophore production, and ACC deaminase activity. Investigating the interactions between the isolated endophytes and C. virgata to understand how these microbes contribute to plant growth, stress tolerance, and disease resistance. Performing field trials to assess the effectiveness of these endophytic bacteria will further enhance the potential application in sustainable agriculture.
-
The authors confirm contribution to the paper as follows: Lata R has conducted all the experiments, data collection, draft manuscript preparation under guidance of Gond SK; Chowdhury S has analysed and participated in the interpretation of results. All authors reviewed the results and approved the final version of the manuscript.
-
All data generated or analyzed during this study are included in this published article and its supplementary information (Supplemental File S1).
Authors would like to thank Department of Botany, MMV, BHU, for providing the necessary facilities. RL is thankful to the UGC, New Delhi for financial support as JRF and SRF. We are thankful to IoE, BHU for providing financial assistance in form of incentive and equipment grant. SKG wants to acknowledge DST-SERB (Govt. of India) for financial aid (EEQ/2016/000555 and EEQ/2020/000485).
-
The authors declare that they have no conflict of interest.
- Supplemental File 1 Figures and tables for the experiments conducted.
- Supplemental File S1
- Copyright: © 2024 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Lata R, Chowdhury S, Gond SK. 2024. Functional characterization of endophytic bacteria isolated from feather grass (Chloris virgata Sw.). Grass Research 4: e006 doi: 10.48130/grares-0024-0005
Functional characterization of endophytic bacteria isolated from feather grass (Chloris virgata Sw.)
- Received: 04 January 2024
- Revised: 11 March 2024
- Accepted: 15 March 2024
- Published online: 28 March 2024
Abstract: The objective of this research was to isolate, identify, conduct biochemical characterization, and evaluate the plant growth-promoting (PGP) characteristics of endophytic bacteria obtained from various parts of feather grass collected from waterlogged rice fields. A total of 43 bacterial isolates from roots, rhizome and leaf of Chloris virgata were isolated. They were subjected to biochemical characterization (Catalase, Oxidase, Indole, Methyl red and Vogues Prausker, oxidative fermentation test). The isolates were tested qualitatively and quantitatively for PGP traits including indole acetic acid (> 0.2·µg mL−1), gibberellic acid production (> 21.03 µg·mL−1), inorganic phosphate solubilization (PSI), siderophore production (> 26.5 PSU), ammonia production (> 120.0 µg·mL−1) and HCN production. The isolates were also subjected to salt (NaCl) and osmotic stress (PEG-6000). Endophytic bacteria were morphologically grouped and selected on the basis of their PGP traits for identification via 16s rRNA sequencing. The identified isolates belonged to two genera namely Bacillus and Glutamicibacter with the former being dominant in the plant. A total of 30 endophytic bacterial isolates identified by 16s rRNA gene sequencing were found to be of Bacillus spp. (25 isolates) and Glutamicibacter spp. (five isolates).
-
Key words:
- Endophyte /
- Plant growth promoting traits /
- Salt stress /
- Halophyte /
- Chloris virgata