-
Rice is an important and staple food crop in the world. Rice seeds not only provide starch, protein, lipids, and mineral nutrients to humans but are also the food source for more than half of the world's population[1]. These carbon compounds accumulated and stored by seeds are increasingly applied in health and medicine fields[1,2]. Furthermore, the research on seed biology can not only improve life quality but also provide guidance for the stable development of ecological environments. Statistics showed that since the last century, the world has lost 75% of agricultural biodiversity with the development of industrial modernization. In recent years, the establishment of 'seed banks' around the world have helped store the genetic information of millions of species and reduced the risk of anthropogenic extinction of biodiversity by 90%, providing important information for the development and utilization of biological resources while protecting the earth's biodiversity[3,4]. Additionally, the innovation of germplasm resources is the strategic goal and the fundamental core of agricultural research. It is the basis for ensuring national food security, promoting long-term sustainable development of agriculture, and the supply of important agricultural products[5].
The mature seed of angiosperms consists of the embryo, endosperm and seed coat. The embryo and endosperm originate from the fertilized egg cell and the central cell, respectively, while the seed coat originates from the periderm of the sporophyte. Seed formation allows the young sporangium and ovule to be protected by the mother and to be well nourished like a mammalian fetus. Seeds also have many structures for dispersal or resistance to adverse conditions, creating favorable conditions for the continuation of plant species. Therefore, in the phylogeny of plants, seed plants can take the place of ferns[6]. Unlike other plant groups, angiosperms have evolved a unique mode of reproduction by double fertilization, whereby the two sperm cells in the pollen of the male gametophyte fuse with the egg cell and the central cell of the female gametophyte, respectively, and further develop into the embryo and endosperm. The emergence of double fertilization in angiosperms have led to the production of endosperm, which is able to provide the necessary nutrients to the newborn embryo to ensure its proper development, laying the groundwork for the reproduction and flourishing of the offspring[7]. Double fertilization changes the nature of seed development and gives the seed of angiosperm plants a more complex structure. In addition to direct maternal-embryonic transport of nutrients from the ectoplasmic interface connecting the base of the embryo stalk to the maternal ectoplasm[8], trans-endosperm transport is also possible, with endosperm cellularization producing an ectoplasmic domain connecting the embryo to the testa, where nutrients will be taken up by the endosperm at the interface between the endosperm and the perithecium, diffusing into the endosperm, importing endosperm cytoplasm from the maternal tissues and then exporting it from the endosperm and taking up nutrients by the embryo[9]. From a developmental point of view, the incipient stages of development of the persistent cereal endosperm resemble those of Arabidopsis thaliana, from the nuclear division of the triploid central nucleus after fertilization of the embryo sac to continued cellularization to form cell walls between the nuclei of individual cells[10]. This difference begins at a later stage, with the differentiation of various tissue types, including the dextran, starchy endosperm, and the rapid accumulation of storage products in the cereal endosperms[11].
Understanding the molecular regulation mechanisms of key traits in rice seed biology is important for improving and breeding superior varieties. Although traditional breeding methods still have the potential to increase yields, improving breeding techniques and efficiency has become a pressing concern due to the constantly rising demand for foods. Therefore, the study of molecular mechanisms that regulate seed development to provide insights for crop breeding has become a mainstream trend for crop genetic improvement. Seed growth and development are complex and precise processes regulated by numerous signal pathways. The G-protein signaling pathway, the mitogen-activated protein kinase (MAPK) signaling pathway, the ubiquitin-proteasome pathway, the BR signaling pathway, and several other signaling have been demonstrated to be related to the regulation[12]. The growth and development of plants and animals cannot be separated by signaling exchanges from intra- and extracellular environments. Heterotrimeric G protein is an important 'signaling switch' in all eukaryotes that is composed of α, β, and γ subunits[13−15]. G proteins complete the transition from the inactivated state to the activated state through the exchange of GDP into GTP binding of the Gα subunit. In the resting state (Gα-GDP form), the GDP-binding Gα forms a complex with Gβγ dimer and anchors to the cytomembrane via its two glycines at the N-terminal. Upon stimulation by upstream signals, GTP replaces GDP to bind to the Gα subunit and activate the protein complex. This leads to the depolymerization of the Gβγ dimer and the Gα subunit, allowing for signal transmission to their downstream effectors, respectively, and regulating various biological processes[13,16]. Animal heterotrimeric G protein pathway is important and well understood to perform a variety of different functions by sensing extracellular signals through cell membrane receptors and transmitting them to ion channels, enzymes, or other effectors[17]. In mammals, G protein-coupled receptors (GPCRs) and G protein-regulating proteins (RGSs) are the key regulating components of the G protein pathway[18]. GPCRs are a kind of receptor protein with a seven-transmembrane structure that senses different signals and promotes the exchange of GTP with GDP to activate the G proteins[19]. The ligand-bound GPCR leads to structural changes in heterotrimeric G proteins, facilitating the exchange of GDP and GTP, and depolymerization of the GTP-bound Gα and Gβγ to transmit signals through downstream effectors, and RGSs primarily function to assist the Gα subunit to hydrolyze GTP, enabling the Gα and Gβγ dimer to reassemble into the resting state[17,20]. Studies on the biological roles of Gα and Gβ in animals indicate that if the phenotypic changes in Gα and Gβ mutants follow a consistent trend, the Gα subunit acts as the signal transmitter, whereas if the trend is opposite, the signal is mediated by the Gβγ subunit. Therefore, there are three different conclusions regarding the roles of the subunits of heterotrimeric G proteins in animals[20−22]: first, the signals are transmitted mainly by the activated Gα subunit, whereas the function of Gβγ is to block its function; second, the signals are transmitted by the activated Gα subunit and Gβγ subunit.; third, the signals are transmitted by the activated Gβγ subunits, while the function of Gα is to block their function. This has accumulated a lot of experience for the study of the G protein signaling pathway in plants; even so, there are still striking differences in G protein signaling pathways between plants and animals.
In animals, ligands bind to GPCRs to promote the formation of Gα-GTP, and GTP binding is the rate-limiting step in G protein signaling. By contrast, in plants, Gα can spontaneously release GDP and form the activated state (Gα-GTP) due to the strong GTP-binding ability, and the trimer is depolymerized, but so far, no typical GPCRs have been identified in plants[17,23,24]. The typical subunits of the G protein have many interacting proteins and their structure is similar to that of GPCRs in plants, but there is no evidence that these proteins facilitate the exchange of GDP and GTP[16,24−27]. Although the members of the G-protein pathway and the structure of the subunits are similar between plants and animals, the number of subunits and the cycle contrast sharply. There are 23 Gα subunits, five Gβ subunits, 12 Gγ subunits, 37 RGSs, and ~ 800 GPCRs present in animals. However, the rice just has one known canonical Gα subunit (RGA1), one Gβ subunit (RGB1), and five Gγ subunits (GS3, DEP1, RGG1, RGG2, and GGC2). Whether there are other novel members of the G proteins in plants is unknown. The RGS box and the 7TM domain of the 7TM-RGS genes are highly conserved in land plants, suggesting the coincidental evolution of these two domains in plants. The differences in intrinsic properties of the Gα subunit in liverworts suggest that the intrinsic regulatory features of the Gα subunit are determined by the binding protein, and the stronger ability of the Gα subunit to hydrolyze GTP makes up for the absence of the 7TM-RGS protein[17]. The absence of RGS protein in rice increases the chances of discovering another mechanism of G protein activation. Animal G proteins function to activate adenylate cyclase and other effector proteins; however, the plant genome seems to have no typical Gα effector reported in animals. Animal G proteins are controlled by a variety of regulatory factors, and no regulatory protein has been identified for rice G proteins; however, the strong GTP-binding ability of RGA1 further suggests that it may be controlled by unknown regulators.
In rice, G protein plays a crucial role in regulating plant height, spike shape, grain size, biotic and abiotic stress responses, nitrogen use efficiency, and responses to almost all phytohormones[28−30]. Additionally, some alleles of atypical Gγ subunits have been widely utilized in breeding[31,32]. Consequently, research focusing on G proteins in rice has been gaining increased attention. As a monocot model plant, rice is an excellent candidate for scientific investigations in plants. Understanding the roles of rice G proteins and their regulatory network has substantial implications for both the basic research of G protein signaling and the genetic improvement of rice and other crops.
-
RGA1 (RICE G PROTEIN ALPHA 1) encodes a typical Gα subunit in rice and exhibits a considerable positive regulatory effect on growth and development[33]. RGA1 was initially identified and cloned from Oryza sativa L. IR-36 by using the cDNA of GPA1 (Gα of Arabidopsis) as a probe[34]. The protein sequence of RGA1 shares 77% identity with GPA1, 86% identity with TGA1 (Gα of tomato), and 42% to 69% identity with mammalian Gα subunits, respectively. It contains a Ras-like domain with the GDP/GTP nucleotide-binding site at the N-terminus common to all Gα subunits, but the potential receptor binding region at the C-terminus exhibits lower similarity to other species, suggesting a distinct activating mechanism of Gα in rice[35]. The RGA1 protein could be detected in all organs, and the protein amount in developing organs was significantly higher than that in developed organs[36]. Besides, the distribution of RGA1 mRNA was similar to that of tubulin[35]. These results imply that RGA1 is constitutively expressed in rice and the expression is regulated based on various developmental stages.
The typical phenotypes of the rga1 mutant are dwarfism, erect panicles, and small round seeds[33]. The D1 gene was mapped on chromosome 5 in the identification of rice gibberellin-insensitive dwarf mutants. Its recessive mutant (d1) showed the same phenotypes as the rga1 mutant, and the amino acid alignment revealed that D1 encodes the α subunit of the G protein[37,38]. Dwarf plants are important materials for studying plant growth and development, and dwarfism is also a desirable trait in breeding[38]. Recently, many alleles of RGA1 were reported and the mutation types included base substitution, base insertion, and base deletion[37,39]. In CM1361–1, a 19 bp insertion between nucleotide positions 354 and 355 in the cDNA of RGA1 led to the lack of three GTP-binding regions, three effector-binding regions, and the receptor-binding region. In DKT-2, the base deletion at positions 932 to 979 caused the loss of the GTP-binding region. In ID-1, two base absence was observed at positions 1003 and 1004, resulting in a frameshift mutation[36]. In RGA1-FH, the function of RGA1 was lost due to a variant of the A-to-T splicing site. They all exhibit dwarfism and produce small round seeds. A sequence polymorphism in the promoter of RGA1 resulted in a low transcript of RGA1, resulting in a semi-dwarf phenotype in cultivar Xueheaizao[39]. The epigenetic mutant Epi-d1 is often a chimeric that shows varying features that range from completely defective seeds to completely normal seeds in one plant, and so does the plant architecture[40]. These allelic mutants show some breeding potential in rice.
RGA1 controls grain size and plant height by balancing cell division and cell expansion[29]. In Nipponbare, the cell length in the lemma of the rga1 mutant was reduced by 32%, while the cell number is only half of that in the wild type. Additionally, the cell number in the fourth leaf sheath, the third internode and the crown root was decreased by 50%, over 50% and 31%, respectively[36]. Furthermore, in indica cultivar M804, the dysfunction of Gα subunit mutant dwarf89 (an A-to-G substitution causing a shorter alpha helix and leading to the deactivation of the Gα subunit) also simultaneously accompanied the decrease in cell number and the increase in cell length in the internode[41]. These findings indicate that the decrease in cell number caused the smaller organs in rga1 mutants.
RGA1 also plays an important role in the regulation of nitrogen transport and nitrogen use efficiency, which are determining factors for grain shape, grain yield, and plant architecture[42]. The nitrogen content of the rga1 mutant in grains was considerably lower than that of the control plant, while it was the opposite in leaves and stem. Furthermore, the rga1 mutant showed lower nitrate-dose sensitivity compared to the wild type in terms of grain yield. And the DEP1-mediated nitrogen response in plant growth was suppressed by the nonfunctioning RGA1[43−45]. In addition, RGA1 is connected to both the GA signaling pathway and the BR signaling pathway[30,37,46]. In the d1 mutant, the activity of α-amylase in the aleurone layer was significantly decreased, and GA-induced genes Ramy1A and OsGAMYB also marked down-regulation. Additionally, the expression of GA-induced Ca2+-ATPase did not increase with GA3 treatment compared to the wild type. Furthermore, higher concentrations of exogenous GA3 were required for internode elongation compared to the wild type[30,38,46]. However, there was no significant difference in the sensitivity of the lower part and the second leaf sheath to GA between the mutant and wild type, indicating that the tissue-specific sensitivity to GA signal mediated by the Gα subunit in rice[30,46]. Two models have been raised to explain the relationship between the G protein pathway and the GA signal in rice[30]: (1) A high-sensitivity GA signaling pathway based on the Gα subunit and its coupled receptors (potentially), and a low-sensitivity GA signaling pathway entirely independent of the G protein pathway. Thus, the loss of RGA1 eliminates the high sensitivity to GA but retains the low sensitivity to GA. This model indicates that the Gα subunit only needs to directly respond to the GA signal to regulate seed germination; (2) The Gα subunit does not transmit GA signaling but regulates the sensitivity of rice seeds to GA. Therefore, knocking out RGA1 could also reduce the sensitivity of rice seeds to GA. This model suggests that RGA1 indirectly participates in the GA signaling pathway through the assistance of other proteins. Otherwise, in response to BR, the transcript level of RGA1 was down-regulated by 24-epiBL in a dose-dependent manner[47]. Additionally, RGA1 interacts directly with the U-Box E3 ubiquitin ligase TUD1 and mediates the early BR response to regulate rice growth and grain size through an unknown mechanism[48].
The Gα metastable epigenetic mutant named d89 was crossed with the rice cultivar 9311, and some semi-dwarf materials without observed adverse traits (setting rate, tillering, and plant height) were bred in an F2 segregating population[41]. The transgene plants containing d1-w (a weak allele of RGA1) exhibited semi-dwarfing, stronger stem, and photosynthesis, higher grain yield and quality, and increased resistance to drought and disease compared to the control plants[49]. These studies provide valuable insights into the genetic variations and the utility of RGA1 in rice production.
-
The rice genome harbors one Gβ subunit known as RGB1 (RICE G PROTEIN BEITA 1), which was originally isolated in 1996[50]. RGB1 is constitutively expressed and functions in a developmental stage-dependent manner; it exhibits a 76% amino acid homology with AGB1 (the Gβ subunit of Arabidopsis), 94% with ZGB1 (the Gβ subunit of Zea mays), and about 40% with the Gβ subunit of animals[50,51].
Knockout of RGB1 results in seedling lethality, while the knockdown lines exhibit smaller seeds, shorter plants, brown internodes, and lamina joint regions[28,51−53]. RGB1 regulates the expression of the IAA synthesis transcription factor OsYUC11 by the mediator OsNF-YB1 through an unknown mechanism. Ultimately, IAA regulates grain filling by promoting starch synthesis and sucrose metabolism in endosperm, thereby influencing grain size[52]. Moreover, in low light conditions, decreasing the expression of RGB1 impacts grain filling by lowering the activity of sink-filling enzymes, including SS and AGPase[53]. Additionally, the effect of GS3-1 RNAi, overexpressing DEP1 and GGC2 mutants on increasing grain length were entirely lost when suppressing the expression of RGB1[54]. Recently, a new QTL SGW5 (suppressor of gw5) with a similar conserved domain and subcellular localization to RGB1 has been reported for grain width regulation and it also interacts with Gα and Gγ subunits, implying it may be a new Gβ subunit in rice[55]. All these findings indicate that RGB1 is a key factor in controlling grain size.
In the RGB1 RNAi plant, the cell length of mortar cells in the inner epidermal tissue does not significantly differ from the wild type, suggesting that the dwarfism of the rice plant is caused by suppressing the expression of RGB1 is attributed to the reduction of cell proliferation[56,57]. RGB1 is necessary for maintaining the homeostasis of the immune system to ensure the seedling development during early germination in rice[51]. The embryo radicle of rgb1 mutant undergoes cell death on 0, 1, 2, 3, and 4 d after germination. Furthermore, the marked upregulation of the immunity marker genes OsPR1a and OsPR10a indicates that the plant immune system may have been overactivated. In maize, ZmGB1 affects kernel row number, and crossing to the tropical line ML103 can rescue the seedling death caused by knockout of the Gβ subunit[58]. The lethality-suppressed mutants showed dwarfism, wider stems, larger SAMs and fasciated IMs, and weaker autoimmunity compared with the wild type. SAM is the source of plant organ formation[59]. These findings not only enrich the regulatory network of the Gβ subunit but also provide a ground-breaking idea for its application in crop genetic improvement.
-
In rice, there are two typical Gγ subunits, RGG1 and RGG2, which negatively regulates seed size[54].There are also three atypical G proteins γ subunits GS3, DEP1, and GGC2, which have different functions for seed development due to the variation of C-terminal, depending on RGA1 and RGB1[60].
The typical Gγ subunit RGG1 in rice has a nuclear localization signal (NLS) at the N-terminus, a GGL domain, and a CaaX isoprenylation motif at the C-terminal, characteristic features of typical type-A Gγ proteins. RGG1 influences endogenous cytokinin accumulation and responses, thereby negatively regulating grain length and panicle length by controlling cell division[61]. RGG2 is a type-B Gγ subunit that functions as a dimer with RGB1 to regulate cell expansion. This regulation is achieved through the mediation of endogenous gibberellic acid (GA) biosynthesis and involvement in GA signaling pathways, ultimately leading to a negative regulation of grain size, organ size, and yield in rice[62]. Noteworthy is the observation that the RGG2 protein is also detected in the plasma membrane, cytoplasm, and nucleus.
The atypical Gγ protein (GGC2) located on chromosome 8 showed 66% and 48% identity with DEP1 and GS3, respectively[54]. GGC2 serves as a positive regulator of grain length and functioned additively with DEP1 in rice.
The DENSE AND ERECT PANICLE 1 (DEP1/EP/qPE9-1/qNGR9) gene encodes a non-canonical Gγ subunit, influencing various growth and development processes in plants. DEP1 was initially cloned on chromosome 9 for erect panicle trait by using a map-based cloning approach and subsequently was named as DEP1[63,64]. The EP, qPE9-1 and qNGR9 were likewise pinpointed at the identical genomic locus[42,64]. DEP1 encodes a novel PEBP (phosphatidylethanolamine-binding protein)-like domain protein and shares similar homology with the N-terminus of GS3. This is followed by a predicted transmembrane domain, two von Willebrand factor C (VWFC) domains, and a VWFC domain at the C-terminus[64,65]. The dep1 variant represents a dominant gain-of-function allele, characterized by the substitution of a 637-bp segment with a 12-bp sequence within exon 5, as compared to the wild-type DEP1. This mutation leads to a 234 amino acid truncation at the C-terminus, eliminating the last two VWFC domains, thereby enhancing grain yield, increasing grains per panicle, and improving nitrogen uptake efficiency[66].
As a Gγ subunit, DEP1 interacts with RGB1 (Gβ) via the GGL (G protein γ-like) domain to form a Gβγ dimer. dep1 loses the TNFR cysteine-rich domain, lifting its inhibition on the Gγ-like domain at the N-terminus, thereby enhancing Gβγ signal transduction. This signal orchestrates a multifaceted role within the plant's developmental trajectory, orchestrating the intricate balance between suppressing longitudinal cell division and plant height during the vegetative phase and concurrently facilitating cell proliferation and panicle branching in the reproductive stage. The resulting increase in meristematic activity leads to shorter inflorescence internodes. Consequently, the dep1 allele produces erect panicle structure, resilient vascular bundles and greater grain number per panicle, and eventually, higher grain yield[66]. Analysis of genetic diversity uncovered a G/C SNP localized within the promoter region of DEP1, inducing a fundamental alteration in a sit II transcriptional regulatory element's core sequence. This alteration exhibits a substantial correlation with both the count of primary and secondary branches as well as the number of grains per panicle[67].
Expressing various truncated DEP1/qPE9-1 by using CRISPR/Cas9 in rice elucidated that the GGL domain located at the N-terminal segment exerts a negative modulatory influence on both rice grain length and grain weight. Nevertheless, this inhibitory effect can be mitigated by the presence of two or three VWFC domains at the C-terminal. Consequently, the C-terminal truncated dep1 protein significantly increases grain number and yield, while reducing grain size and grain weight. Hence, DEP1 thus emerges as a pivotal facilitator of grain length and weight in rice[68,69]. Primarily, DEP1/qPE9-1 enhances grain size by promoting endosperm cell proliferation. In the grain size model involving Gγ proteins, the pairing of DEP1 or GGC2 with RGB1 enhances grain size through tail-mediated signaling. Conversely, GS3 diminishes grain size by obstructing the DEP1 and GGC2 interaction with RGB1. The tail-mediated autodigestion of GS3 within the RGB1-GS3 complex establishes a dynamic equilibrium between blocking and enabling the interaction of DEP1/GGC2-RGB1. An absence of the GS3 tail leads to its accumulation and subsequent occupancy of a substantial amount of RGB1, resulting in shorter grains[51,70].
DEP1 directly interacts with the conserved keratin-like domain found in MADS transcription factors, thereby acting as cofactors to amplify the transcriptional activity of OsMADS1. This interaction promotes the cooperative transactivation of shared target genes, thereby orchestrating gene expression patterns associated with grain size and morphology. And the VWFC domain of DEP1 is essential for this DEP1-OsMADS1 interaction[70]. IDEAL PLANT ARCHITECTURE1 (IPA1) plays a crucial role in regulating rice plant architecture and significantly bolstering grain yield. By directly binding to the GTAC motifs present within various loci of the DEP1 promoter, IPA1 acts as a positive modulator of DEP1 in controlling plant height and panicle length in rice[71]. GL7 (Grain Length on Chromosome 7) encodes a rice homolog of the LONGIFOLIA (LNG) protein of Arabidopsis thaliana, working alongside TONNEAU 1 (TON1) and protein phosphatase 2A (PP2A) holoenzyme (with TON2/FASS as the regulatory subunit) to form a TON1-TRM-PP2A complex (TTP complex)[72]. These components play a critical role in modulating the microtubule array to regulate cell growth and division. Both DEP1 and dep1 variants can interact with TTP complex components, such as OsTON1b and OsTON2, thereby influencing grain longitudinal elongation by inhibiting the TTP complex in microtubule organizing centers[73]. GR5 encodes an AP2-type transcription factor featuring an AP2/ERF domain, which controls grain size by regulating the expression of genes for grain size determination and cell cycle that includes DEP2 (DENSE AND ERECT PANICLE2), DEP3 (DENSE AND ERECT PANICLE3), DRW1 (DWARF-RELATED WD40 PROTEIN1), and CyCD5;2 (LOC_Os12g39830). DEP1 positively modulates the transcriptional activity of GR5, while GGC2 has a negative effect on GR5[74].
DEP1 and dep1-type accessions show considerable genetic variation in plant height and tiller numbers across diverse nitrogen fertilization gradients. The GGL domain of DEP1 establishes interactions with RGB1 at both the cytomembrane and intranuclear levels. DEP1 and RGA1 collaborate with a shared signaling cascade governing a prototypical nitrogen-induced growth response, with the VWFC domain of DEP1. Enhanced RGB1 activity, however, inhibits the nitrogen response[42]. Recently studies revealed that under both limited and sufficient nitrogen supply, dep1 improves grain yield and nitrogen use efficiency (NUE) by increasing nitrogen and dry matter transport, attributed to higher GS (Glutamine synthetase) activity in leaf[42,64]. The GS activity is essential for nitrogen assimilation and enhances the movement of dry matter and nitrogen from the stem to the spikelet during the grain-filling period. Under limiting nitrogen conditions, near-isogenic lines (NILs) of dep1 outperform NILs of DEP1 in terms of tiller and grain number per panicle[75,76]. The dep1 also reduces the enzyme activity of Rubisco and PEPC. Yield traits like panicle number, spikelet number, grain filling, and grain weight were influenced by qPE9-1/qpe9-1 allele, exhibiting diverse responses to varying nitrogen supply rates in field conditions. Particularly, panicle number emerged as a significant determinant for grain yield between DEP1 and dep1-type cultivars. Nonetheless, regardless of nitrogen supply rates or planting densities, the DEP1-type variety consistently displayed superior grain weight compared to dep1-type cultivars[77].
DEP1/qPE9-1 appears to play a positive role in starch accumulation in seeds largely due to its promotion of gene expression associated with starch biosynthesis during the mid to late grain-filling stage, thus extending the grain-filling duration[69]. A previous study indicated the erect panicle allele qPE9-1 does not affect the eating and cooking qualities of milled grains[78]. However, recent investigations into the textural attributes of parent lines and recombination of inbred lines across four regions suggests that dep1 may alter rice eating quality through the regulation of amylopectin chain length distribution[79]. Besides, some reports suggest that qPE9-1negatively affects plant height, panicle length, tillers per plant, leaf length, grain weight, and overall grain yield per plant[78]. Under the genetic background of indica rice, erect panicle types exhibit significantly lower grain yields than drooping panicle types[80]. This variation suggests that dep1 interacts with other genes in different contexts, because its influence on grain yield is a complex, multi-gene-controlled quantitative trait.
-
GS3 (GRAIN SIZE 3), encoding an atypical Gγ subunit, is a negative master regulator of grain length and size/yield[81,82]. As a significant quantitative trait locus (QTL), GS3 is a key regulator governing both grain weight and grain length in rice. It has relatively minor influences on grain width and thickness[81]. GS3 consists of a tumor necrosis factor receptor/nerve growth factor receptor (TNFR/NGFR) family cysteine-rich domain, a von Willebrand factor type C (VWFC), a transmembrane domain in the C terminus, and a plant-specific organ size regulation (OSR) domain in the N terminus. The OSR domain functions as a negative regulatory motif that is inhibited by the TNFR/NGFR and VWFC domains[81,82].
A premature termination caused by the C-A mutation in the second exon of GS3 fades the effect of the OSR domain, resulting in grain elongation[81,83,84]. This mechanism indicates that the loss function of GS3 generates long grains; otherwise producing short grains. There are six GS3 alleles (GS3-1, -2, -3, -4, -5, -6) having been detected in rice[82,85]. Among these alleles, the C-A mutation mentioned above presents in a long-grain variety, Minghui 63 (GS3-3). Such an allele is also regarded as gs3, clearly in agreement with the recessive nature of the long- grain phenotype. In contrast, Zhenshan 97 (GS3-1) and Nipponbare (GS3-2) with all the predicted domains, exhibit a wild-type configuration, and display an intermediate grain size. The GS3-4, -5 and -6 alleles are represented by Chuan 7, SYB6, and Zhimali, respectively. These alleles carry diverse variants in exon 5, encoding truncated proteins that lack the TNFR/NGFR and VWFC domains[8]. These particular alleles exert the most pronounced effect in decreasing grain length due to the total deficiency of repressive effects on the OSR domain[81,82,84]. GS3 is highly expressed in young panicles, and the signal gradually decreases as panicles develop[82]. Weak signals can be detected in other tested tissues, including embryo, shoot apical meristem, leaf, and stem. Interestingly, GS3 is also highly expressed in root tips[82]. This special expression pattern may indicate various functions of GS3, such as alkaline stress response[86]. At the cellular level, GS3 regulates grain length by modulating the number of cells within the upper epidermis of the glume. This phenomenon suggests the involvement of GS3 in the regulation of cell division[84].
GS3 interacts with RGB1, forming a Gβγ dimer to further regulate grain size. Significantly, the presence of DEP1 and GGC2 in complex with RGB1, leads to enlarged grain length. However, GS3 counteracts this effect by competitively interacting with RGB1. It has been further pointed out that GS3 encoded by GS3-4/-5/-6 with shorter C-terminal tails produce shorter grains. A tail-mediated self-degradation can explain this phenomenon[87]. Chang Li Geng 1 (CLG1), which encodes an E3 ligase. CLG1 interacts and ubiquitinates the full-length GS3-1 and -2 in both the cytoplasm and cytomembrane, while the ubiquitination of truncated GS3-4 (lacking the Cys-rich tail) only occurs in the cytomembrane. Therefore, GS3-1 and -2 are led to lysosome for degradation, yet the ubiquitinated GS3-4 cannot be sorted to lysosome, only to anchor on the membrane, continually occupy RGB1, resulting in shorter grain length[87]. This pathway has further filled the relationship network of G-proteins in plants, and highlighted the significance of the C-terminal tail related to protein degradation[54,64,87]. GS3 and DEP1 combine with RGB1 to form a G dimer, which has the potential to translocate into the nucleus for the subsequent regulation of transcription factor activities. For instance, GS3 and dep1-1 can combine with a MADS transcription factor (encoded by qLGY3), enhancing OsMADS1 transcriptional activity and promoting the cooperative transactivation of common target genes, thereby regulating grain size and shape[69].
The genetic relationship between GS3 and another three seed-size related genes (GW2, qSW5/GW5 and GIF1) has been proven genetically[88]. GW2 and qSW5 up-regulate the expression of GS3, while GIF1 is negatively regulated by GW2 and GS3. In addition, the effect of qSW5 on seed length is inhibited in the presence of GS3 alleles and the effect of GS3 on seed width is inhibited by the qSW5 allele[88]. gs3 and qgl3 additively regulate rice grain length[89]. Some researchers have pointed out that the regulation of grain length by qGL3 and GS3 may be involved in the BR signaling, and other signal pathways that regulate cell growth and cell cycle[89]. BR signaling pathway plays an important role in manipulating the growth of plants as well as adaption to stress[90,91], which exactly corresponds with the effect of GS3.
GS3 has been considered as a conserved protein in regulating seed size in plant systems. TaGS-D1 and TaGS3 in wheat, ZmGS3 in maize, GC1/AT1 in sorghum and AGG3 in dicotyledonous plants Arabidopsis, are homologous proteins of GS3[86,92−95]. However, the effects of GS3 homologs in Cruciferae is opposite to the effect in cereals[96]. This difference is relative to alternative splicing, a general mechanism that regulates gene expression at the post-transcriptional level[97]. In cereals, there are two dominant isoforms of GS3, GS3.1 and GS3.2. GS3.1 encodes the full-length protein, resulting in decreased grain size. Meanwhile, GS3.2 generates a truncated protein only containing an OSR domain, showing no significant effect on grain size. GS3.2 disrupts GS3.1 signaling by competitively interacting with RGB1. Therefore, the alternative splicing mechanism inhibits the negative effect of GS3.1 on grain size regulation[97]. Alternative splicing is not detected in the GS3 homologs of Cruciferae[97].
As an important gene participating in plant growth and grain size, GS3 has wide-ranging applications in increasing rice production and yield. There are many examples of improving rice variety via updating the GS3 locus. Both grain length and weight per plant of the improved line that was introgressed by a ~117-kb segment including the gs3 allele from the donor GKBR, having an impressive raise, compared with the elite japonica variety of Kongyu 131[31]. The gs3 allele from Akita 63, enlarges grain size, improves nitrogen-use efficiency (NUE), and thus increasing the rice yields. When transferred into normal-grain japonica cultivar, Notohikari, this allele largely confers the superior trait with a larger grain size and high NUE[32]. gs3 is also a key to raising improved crops for saline and alkaline soils. KY(gs3), Kongyu 131 with an introduced gs3, shows a higher alkaline tolerance than Kongyu 131, thereby increasing rice production in highly sodic areas[89]. Furthermore, the gs3 allele decreases methane emissions from methanogens by diverting a larger proportion of photosynthates to the grain and reducing the allocation to the root. Milyang360 with the gs3 allele can largely reduce methane emissions and nitrogen fertilizer input[98]. Due to its various functions, GS3 has been widely applied to improve crop traits.
-
Besides the functions in seed development, G proteins have many other important functions in rice and other plants. Knockout of RGA1 could abolish the epidermal cell death induced by ethylene and H2O2[99]. Furthermore, RGA1 participates in the strigolactone-mediated regulation of tillering[100]. The rga1 mutant exhibits increased tillering, accompanied by the down-expression of D10 (strigolactones synthetase), which is a negative regulator of tillering. Under water scarcity conditions, rga1 mutant exhibited increased survival capacity due to the lower leaf temperature with a smaller driving force for water loss compared to the wild type, which could guarantee longer photosynthesis[101]. Meanwhile, RGA1 exerts a negative regulatory effect on thermo-tolerance by impacting carbohydrate metabolism and the energy supply in rice[102].
RGB1 is also crucial in disease resistance, stress resistance and environmental adaptation[50,52]. There are abundant stress-related cis-regulatory elements found in the promoter region of RGB1 and it was localized on the plasma membrane, cytoplasm, and the nucleus, suggesting its potential interaction with abiotic stress-related transcription factors in nucleus to regulate abiotic stress. Additionally, a significant up-regulation of the RGB1 transcript was detected under stress treatment (cold, K+, Mn2+, and Zn2+), while high temperature and heavy metal (arsenite, arsenate, cadmium, and lead) treatment did not lead to noteworthy changes in RGB1 transcription[51,103,104]. Overexpression of RGB1 could confer higher salt resistance to rice[105]. After high concentrations of salt treatment (120 mm–300 mm NaCl), the chlorophyll content of overexpressing transgenic lines was significantly higher than that of wild-type plants, without obviously cell necrosis on the leaves. Additionally, overexpression of RGB1 could improve the sheath blight disease resistance of rice[106].
Besides the grain size regulation, dep1 could promote the expression of RBOH (Respiratory burst oxidase homolog) to maintain high levels of endogenous H2O2, regulates cortical cell death, and facilitates aerenchyma formation in rice roots[107]. LPA1 (loose plant architecture 1) can bind to the promotor of PIN1a to enhance the planting density and resistance to sheath blight disease[108]. DEP1 interacts with the indeterminate domain (IDD) of LPA1 and inhibits its DNA-binding ability. As a result, the transcription of PIN1a dependent on LPA1 was weakened, leading to a decrease in PIN1a expression. Additionally, the cysteine-rich region of DEP1 contributes to cadmium tolerance in plants[109]. RGB1 has a positive influence on ABA biosynthesis, while qPE9-1, which is regulated by RGB1, serves as a suppressor in ABA-dependent drought-stress responses[69]. GS3 is related to the flag leaf size regulation, stigma exsertion, alkaline sensitivity, high temperature, brown planthopper resistance, photosynthesis allocation, and methane emission[98,110−113].
-
In summary, plant heterotrimer G proteins are highly conserved and have similar functions, contributing to yield, apical meristem activity, resistance, and nutritional efficiency (Fig. 1). The signaling mechanism of the heterotrimer G protein in plants and animals is fundamentally different, the relationship of G protein and small G protein is not clear, and it is urgent to study heterotrimer G protein signal conduction in plants, especially model crop rice: it not only contains all aspects of the needs of the three major crops industry chain, but also has basic and frontier scientific problems behind the industry chain. As a model plant of monocotyledonous, rice has great advantages in molecular, genetic, biochemical, and other studies. In addition, the research on rice G protein has a good preliminary basis. The study of heterotrimer G protein signaling mechanism in rice can lay a foundation and reference for the study of universality, conserved, and special G protein signaling mechanism in other crops: receptors and ligands may be conserved in heterotrimer G protein signaling in plants, and the research basis is first done (and the progress is also advanced) in the model crop rice, and then verified in other crops. The middle and downstream effectors may be functionally different in the three crops, it is necessary to make parallel studies separately, and it is possible to find functionally different middle and downstream effectors.
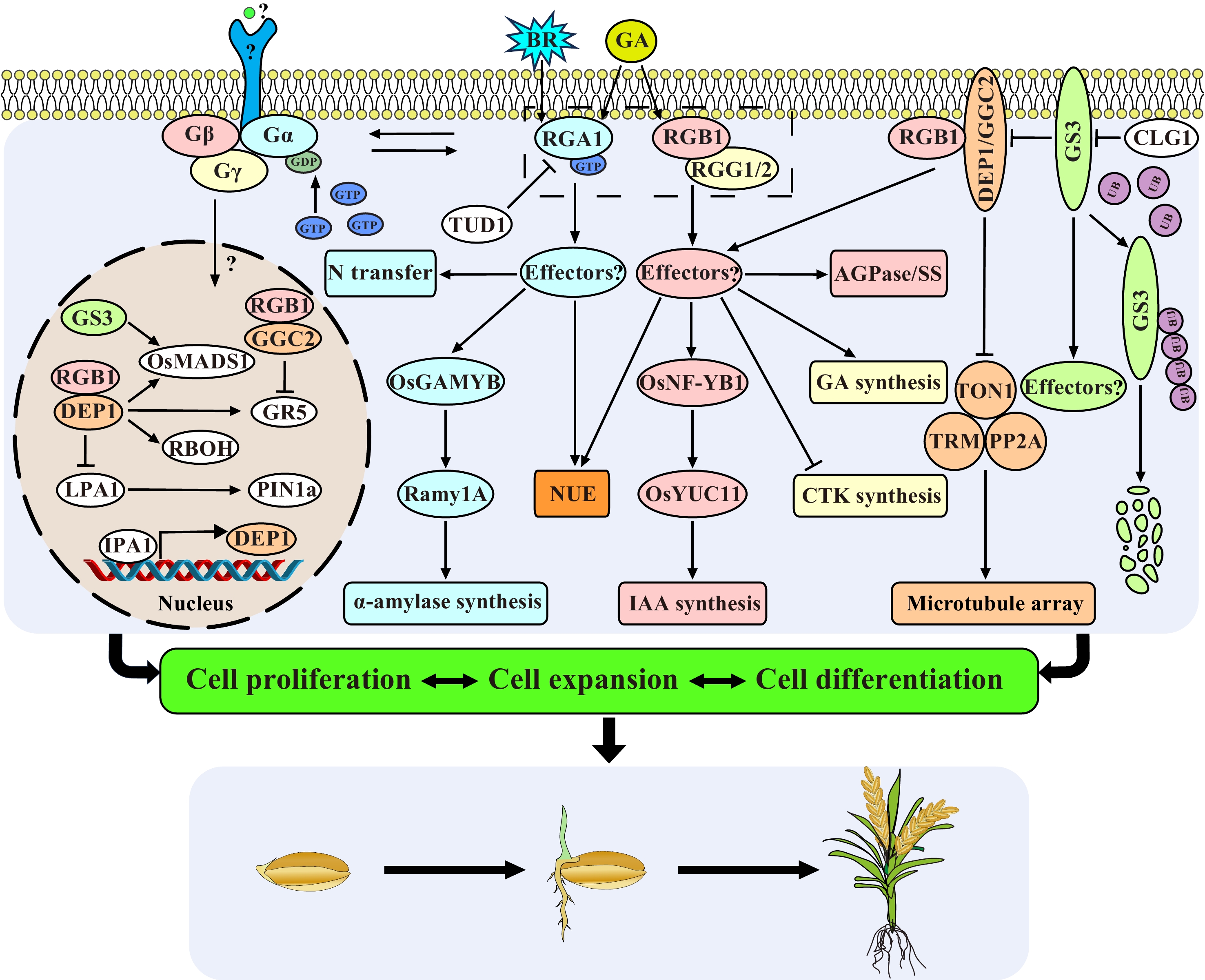
Figure 1.
Regulatory network of G proteins in rice seed growth and development. G proteins transmit the BR, GA and other unknown signals, and regulate the nitrogen transport, the synthesis of plant hormones, the enzyme activity of AGPase, SS and α-amylase related to grain filling through many unknown downstream effectors, thereby control grain size and development. The β and γ subunits can enter into nucleus to influence the transcriptional activity of several transcription factors to affect cell proliferation and cell expansion. RGA1 interacts with TUD1 and mediates the early BR response. DEP1 influences grain longitudinal elongation by inhibiting the TON1-TRM-PP2A complex. CLG1 acts as the upstream of GS3 and promote its degradation. The external signals, the direct upstream receptors, and direct downstream effectors of G proteins are unknown.
As a signal transfer hub, sufficient evidence has shown that rice G protein plays an important role in seed germination, growth and development, and gain yield (Fig. 1). Each subunit is pleiotropic, individual, or in combination to affect the entire life cycle of rice. Knockout of RGA1 can lead to extreme dwarfism and smaller seeds, but it also improves the tillering, lodging, and drought resistance. Moreover, RGA1 also participates in some signaling pathways (BR, GA, etc.) to regulate the grain size and grain filling. RGB1 is essential for the survival of rice, as a knockout leads to seedling lethality and down-regulation results in shorter, thinner, and narrower grains, while overexpression of RGB1 enhances the ability to resist salt, cold, and drought stress. RGB1 is also involved in the synthesis of IAA and nitrogen utilization efficiency. RGG1 and RGG2 also contribute to grain size in rice, and may also be involved in the GA synthesis pathway to coordinate cell proliferation. The DEP1 knockout mutant exhibits smaller grains, and the overexpressed plants have observed larger grains, and it also influences the starch synthesis during the filling stage. dep1 could increase the number of primary and secondary branches and the number of grains per panicle of rice, thereby increasing grain yield. And it could also improve the nitrogen utilization efficiency, and increase the lodging and drought stress resistance, though resulting in smaller grain size and poor quality. GS3 is a major QTL controlling grain shape, and knockout of GS3 resulted in a dramatical increase in grain size, however, the number of grains per panicle was reduced.
Gene editing technology has been widely used for crop improvement. The editing of the promotor of Waxy, GWD1 and Xa13 genes could improve the rice quality and resistance to Xanthomonas oryzicoia[114−116]. The function of the G protein subunits are involved in some characteristics of ideal plant/spike shape of rice (compact plant shape, the high effective number of spikes, large grain size, disease resistance, resistance to downfall, etc.), therefore, coordinating the function of G protein subunits by genome editing to aggregate the advantageous traits can provide a new direction for molecular breeding.
DEP1 only occurs in japonica rice and its introduction into indica rice will make indica rice produce high yields and show better traits in indica background, which indicates that indica rice contains unique genes that interact with dep1, which needs to be further studied. On the other hand, dep1 shows different functions in different backgrounds, indicating a complex regulatory network, and now the genes of the dep1 interaction network are gradually being uncovered, and the G protein signaling pathways mediated by dep1 are diverse, involving various aspects of seed biology, which can respond to different needs, varieties, and growing environments. Utilizing the dep1-mediated G protein signaling pathway and interaction proteins can offer significant benefits. dep1 also has the characteristics of lodging resistance, strong light and air flow ability in agricultural traits, and has a wider application prospect. However, further improvements are needed for the 1000-grain weight and quality of DEP1.
Based on current progress, five key scientific questions about the regulation mechanism of the G protein pathway in plants is proposed (Fig. 1): (1) What are the direct upstream signals and receptors of G proteins? G protein signal transduction is studied mostly in the downstream pathway in plants, but its upstream signals and receptors are unknown. Plant-specific receptor-like kinases (RLKs) represent one of the largest gene families in plants and are involved in the perception of various environmental, chemical, and developmental signals. Recently, experiments have demonstrated that plant G proteins interact with plant-specific receptor-like kinases (RLKs)[117]. Signal transduction by endosomal plasma membrane receptors in the nucleus is also an exciting new topic; (2) What are the major downstream effectors of Gα, Gβ, and Gγ signaling for various functions? Future studies aimed at discovering new components and their signaling mechanisms will surely reveal multiple new aspects of the signaling module of the missing G protein components; (3) The rice genome only encodes a single Gα subunit and is anchored to the cell membrane, but it is involved in so many biological processes. How does the Gα subunit achieve precise various signal regulation? (4) Is it really necessary for the Gβ and Gγ subunits to form a heterodimer like animals to function in plants even though their contributions to grain size or other functions are opposite? Can Gγ and Gβ signal independently in plants? (5) How do Gβ and Gγ shuttle from the membrane to the nucleus, and what is the function of the shuttling? In plants, the entry of membrane protein or cytosolic protein into the nucleus is an important signal transduction mechanism, which is involved in plant growth and development, disease resistance, and stress responses[118]. However, the mechanism of how the Gβγ enters the nucleus remains unclear. We thus call for an international coordinated effort in studying and solving these challenging problems in the form of a project named 5QGPS (short for five key scientific questions of G protein signaling in plants).
G proteins are highly conserved and have similar functions in plants, and there are many basic differences between plants and animals in G protein signaling. Rice as a model plant has many important research basis in G proteins, and thus, future progresses of rice G protein signaling can provide solid references and insights for other plants.
-
The authors confirm contribution to the paper as follows: Xiong M finished the abstract, and part of the introduction, Gα, Gβ, and part of perspective. Zhang H finished the Gγ of DEP1, and part of the introduction and perspective. Huang Y finished the Gγ of GS3 and part of the introduction and perspective. Li Y conceived and designed the review and wrote the manuscript.
-
Data sharing not applicable to this article as no datasets were generated or analyzed during the current review paper.
This study is supported by grants from the National Key Research and Development Program of China (2021YFF1000202, 2022YFD1200103), National Natural Science Foundation of China (U22A20470, 32072042), Ten-thousand Talents Programs, Hubei Hongshan Laboratory (2022hszd025, 2021hszd005), the Key Research and Development Program of Hubei (2023BBB135, 2022BBA0033) and the Fundamental Research Funds for the Central Universities (2662023PY002).
-
The authors declare that they have no conflict of interest.
-
# Authors contributed equally: Meng Xiong, Huiying Zhang, Yuxin Huang
- Copyright: © 2024 by the author(s). Published by Maximum Academic Press on behalf of Hainan Yazhou Bay Seed Laboratory. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Xiong M, Zhang H, Huang Y, Li Y. 2024. G protein regulation in rice seed biology. Seed Biology 3: e010 doi: 10.48130/seedbio-0024-0008
G protein regulation in rice seed biology
- Received: 03 February 2024
- Revised: 10 April 2024
- Accepted: 30 April 2024
- Published online: 13 June 2024
Abstract: Seeds are not only the organ for producing progenies but also the vector of grain quality and yield in cereal crops. Research focusing on seed biology to improve crops is important for ensuring food security and addressing hunger issues worldwide. The heterotrimeric G protein consists of Gα, Gβ, and Gγ subunits, which act as a conserved signaling switch to transmit extracellular signals to downstream effectors, thereby regulating various biological functions. G proteins in rice control numerous important agronomic traits, biotic and abiotic stress responses, with significant application potential, thereby drawing increasing attention from researchers and breeders. Here, the recent progress of G protein signaling in seed development, including grain size, number, yield, quality, and stress resistance, is summarized with a focus on the molecular and cellular regulation of the G protein signaling pathway in seed biology, and some understandings in potential applications are offered. Investigating the regulation of the G protein pathway can not only address the critical issues in rice grain development but also offer novel insights into rice breeding. Although it is clear for G protein pathways in mammals, the upstream signals, receptors, and direct downstream effectors of G protein pathways in plants are still unknown in many important biological progresses. Five key scientific questions underlying G protein signaling in plants are therefore proposed to call for an international coordinated effort in studying and solving these problems in the form of a project named 5QGPS (five key scientific questions of G protein signaling in plants).
-
Key words:
- Rice /
- Seed /
- Biology /
- G proteins













