-
Ornamental plants hold considerable economic value. They are exhibit a diverse range of shapes and vibrant colors, with their leaves, flowers, fruits, and stems all possessing ornamental value[1]. These plants are cultivated extensively worldwide to meet market demand. However, compared to model plants, ornamental plants often have lower regeneration efficiency, with some ornamental plants even unable to regenerate using traditional methods, impeding their rapid propagation, virus-free breeding, and molecular breeding[2]. Traditional propagation techniques of ornamental plants, such as bud division, cuttage, and grafting, pose risks of disease transmission, leading to a swift decline in quality, manifested by developmental delays, leaf spotting, poor rooting, color variations in flowers, and flowering abnormalities. Thus, establishing an efficient rapid propagation and in vitro regeneration system for ornamental plants could resolve these quality issues and improve reproduction efficiency, enabling large-scale production[3,4].
For decades, research on the regeneration of ornamental plants has mainly focused on selecting explants, optimizing growth media, employing plant growth regulators (PGRs), and environmental factors[5]. Despite these efforts, breaking through the regeneration bottleneck in some ornamental plants remains a formidable challenge. This challenge restricts the production, breeding, and diversification of high-quality cultivars and varieties. Moreover, many ornamental plants lack a genetic transformation system due to their limited regeneration capacity, which hinders molecular breeding[6].
Recent theories suggest that the regenerative capacity of plants is primarily influenced by the fate of their cells and tissues, which are intricately linked to their original tissue origins. Upon initiation of the regeneration process, plant cells undergo a transformation in fate, leading them to progress into the regenerative phase[7]. This process is intricately influenced by factors such as regeneration-associated genes and epigenetic modifications. Enhanced insight into the regulation of plant regeneration has facilitated the employment of diverse molecular tools to tackle concerns associated with the regeneration of ornamental plants. These tools encompass the substitution of transgenic material screening techniques, utilizing regeneration-associated genes, epigenetic modifications, and utilization of novel genetic modification techniques, among others[8,9].
This paper provides an overview of the traditional research achievements and recent advancements in plant regeneration. It summarizes the multiple factors influencing plant regeneration, including regeneration pathway preferences, traditional ornamental plant regeneration research encounters obstacles, as well as the latest genetic and epigenetic influences on ornamental plant regeneration. Furthermore, it also discusses the prospects of utilizing the CRISPR/dCas9 activation system, monitoring chromatin accessibility, and applying non-tissue culture-based genetic transformation techniques in ornamental plants. These efforts aim to address the challenges faced by ornamental plants during the regeneration and genetic transformation processes[10−13].
-
There are two common regeneration pathways in plants: organogenesis and somatic embryogenesis (Fig. 1). The organogenesis pathway involves a vascular connection with the parent tissue, while the somatic embryogenesis pathway does not involve such a connection[14]. Both organogenesis and somatic embryogenesis pathways have been observed during in vitro regeneration of ornamental plants.
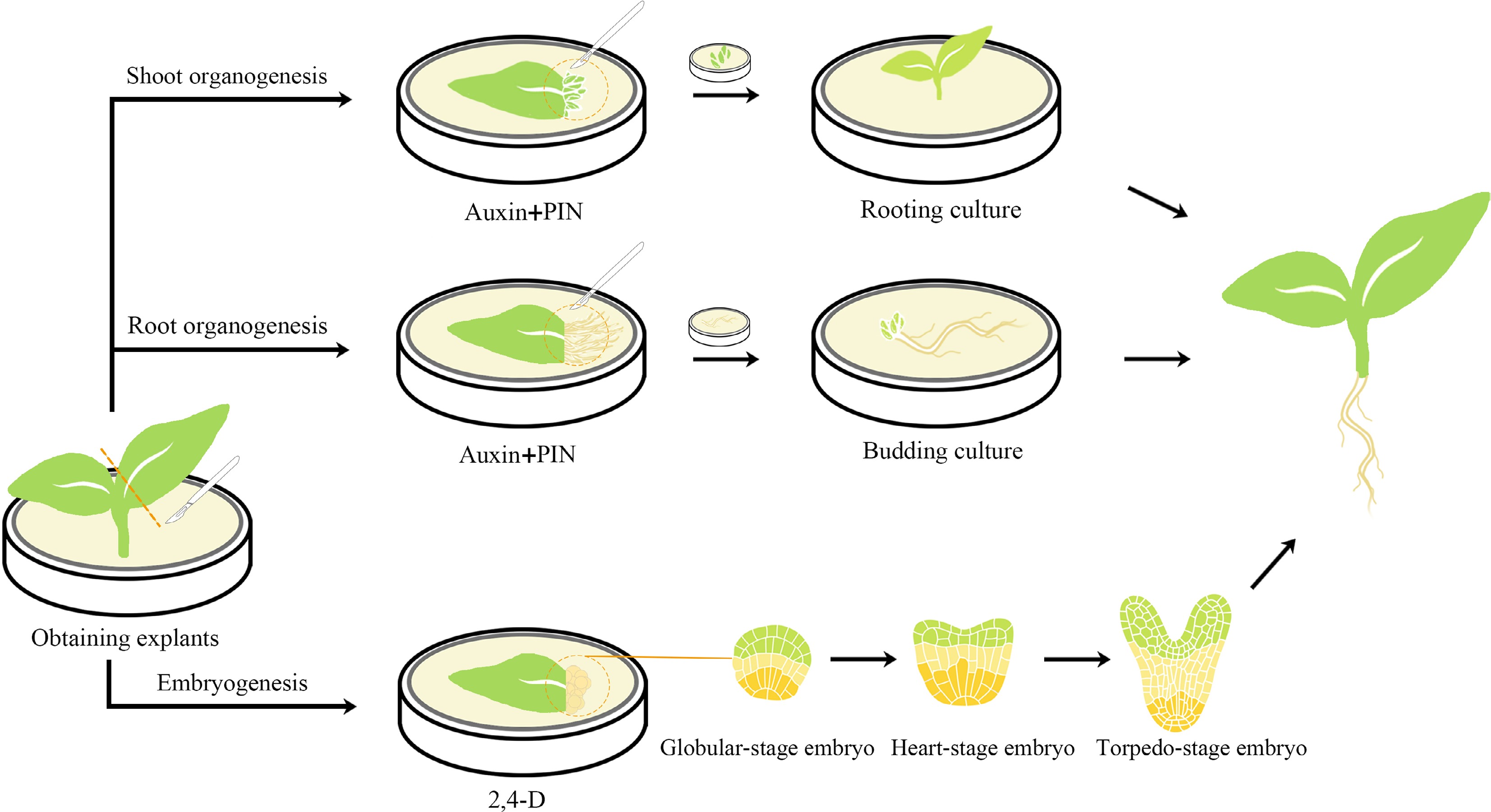
Figure 1.
Schematic diagram of organogenesis and somatic embryogenesis processes in plant regeneration. Organogenesis can be achieved through the induction of bud formation, followed by in vitro culture of the newly formed buds and subsequent cultivation of independent plants through root culture. Alternatively, root formation can be induced, and the roots can be cultured in vitro to stimulate bud formation and generate new individuals. The majority of somatic embryogenesis processes necessitate induction with 2,4-D, and after successful induction, somatic embryos go through several stages before shoot formation.
Different types of ornamental plants show preferences for different pathways of regeneration. In the case of dicotyledonous herbaceous ornamental plants, such as chrysanthemum (Chrysanthemum morifolium)[15], Dianthus chinensis[16] and pyrethrum (Tanacetum cinerariifolium)[17], using stems or leaves as explant induction is the primary method for promoting organogenesis. Another common regeneration strategy involves the initial formation of root organs, followed by root budding, such as in species like oregano (Origanum vulgare)[18] and Taraxacum koksaghyz[19]. Conversely, monocotyledonous ornamentals frequently undergo regeneration through somatic embryogenesis, a process observed in species such as tulips (Tulipa gesneriana)[20], ornamental banana (Smusa spp.)[21], and cut lilies (Lilium longiflorum)[22]. However, certain monocotyledonous ornamental herbaceous plants also undergo regeneration through organogenesis, such as Lilium candidum[23]. It has been observed that dicotyledonous herbaceous ornamentals prefer organogenesis, while monocots show a tendency towards somatic embryogenesis. However, in woody dicotyledonous ornamentals, there is no clear preference for a specific regeneration pathway.
During organogenesis, ornamental plants can be classified into direct regeneration and indirect regeneration based on the presence of callus tissue. Direct regeneration, commonly observed in dicotyledonous ornamentals such as pyrethrum[24] and plane tree (Platanus acerifolia)[25], is typified by the absence of a visible callus during the regeneration process. In this pathway, the procambium within the vascular tube acts as the source of cells for regeneration. However, the limited presence of protocambium in mature leaves of monocotyledonous plants pose challenges for direct regeneration. As such, regeneration in these species often necessitates the induction of a renewable callus from juvenile tissues, which then serves as the primary substrate for indirect regeneration. For instance, the stem of Sansevieria trifasciata has been effectively utilized as explant material to induce callus formation for subsequent regeneration[26]. Overall, the selection of a regeneration pathway in ornamental plants is heavily contingent upon specific plant traits and their developmental stages (Table 1).
Table 1. Regeneration mode of some ornamental plants.
Species Plant category Plant regeneration pathway Ref. Chrysanthemum morifolium Dicotyledonous herbaceous plant Organogenesis [27] Dianthus chinensis Organogenesis [16] Tanacetum cinerariifolium Organogenesis [17] Origanum vulgare Organogenesis [18] Taraxacum koksaghyz Organogenesis [19] Tulipa gesneriana Somatic embryogenesis [20] Smusa spp. Monocotyledonous herbaceous plant Somatic embryogenesis [21] Crinum
malabaricumSomatic embryogenesis [28] Fritillaria meleagris Somatic embryogenesis [29] Lilium longiflorum Somatic embryogenesis [22] Sansevieria trifasciata Organogenesis [26] Lilium candidum Organogenesis [23] Aeschynanthus pulcher Woody plant Organogenesiss [30] Robinia pseudoacacia Organogenesis [31] Koelreuteria paniculata Somatic embryogenesis [32] Cinnamomum camphora Somatic embryogenesis [33] Platanus acerifolia Organogenesis [25] -
In the realm of plant regeneration research, the focus has been on examining both intrinsic and extrinsic factors influencing the regeneration processes of ornamental plants. Intrinsic factors consist of genetic variations, the selection of explants, and their physiological maturity. Whereas primary external factors include the choice of growth media, microbial impact, and growth environmental conditions[34].
The effects of different genotypes, the selection of explants, and the physiological age of explants on the regeneration of ornamental plants
-
Genotypic and species-specific variations significantly impact plant regeneration. Model plants, characterized by their consistent and simple genetic backgrounds, facilitate more controllable and predictable regeneration outcomes. Their genetic tractability allows for the broad application of established regeneration techniques across various genotypes[35]. Conversely, ornamental plants exhibit a higher degree of genetic diversity due to selective breeding for diverse attributes such as color, shape, and size[36]. The diversity within species leads to variations in regenerative capacities among different genotypes, making the regeneration of ornamental plants often more complex compared to model plants.
Variations in regeneration attributed to genotype differences have been observed in several ornamental plant species. For example, studies on Petunia hybrida and Calibrachoa elegans have demonstrated genotype-dependent variations in protoplast cell division and stem regeneration[37]. Similarly, regeneration efficiencies in Sansevieria spp. vary significantly across genotypes under the same culture medium conditions, ranging from 0% to 73.3%[38]. Besides, Paeonia ostii exemplifies varying rates of shoot organogenesis among four different genotypes, ranging from 0% to 20.81% following the induction of somatic embryos[39]. While the genotype significantly influences the regeneration efficiencies, ornamental plants within the same botanical family often share similar regeneration pathways. Taking the Compositae family as an example, organogenesis is a common feature in these plants and the regeneration conditions typically favor higher cytokinin levels over auxin, as seen in species like sunflower (Helianthus annuus)[40], pyrethrum[17], chrysanthemums[27], and calendula (Calendula officinalis)[41].
The choice of explant material is crucial for successful tissue culture. Specific explant organs such as leaves or hypocotyl are commonly used in model plants like Arabidopsis and Nicotiana benavidesii. However, in ornamental plants, a broader range of explant types is employed, depending on the regeneration goal and the plant's characteristics. Various explant types, including leaves, petioles, hypocotyls, cotyledons, embryos, internodes, and roots, significantly impact the outcomes of plant tissue culture[42]. For example, the endosperm explants from Passiflora foetida have demonstrated high efficiencies of adventitious bud regeneration[43]. In the cultivars of 'Brasil', 'Capitola' and 'Jewel Time Yellow' of chrysanthemum, the ovaries demonstrated greater organogenic potential than that of ovules in both callus formation and shoot development[44]. Axillary bud explants of tuberosa (Polianthes tuberosa) displayed higher regeneration efficiency compared to bulblets[45], and in Lilium brownii, scale explants had higher regeneration efficiencies than leaf and petiole explants[46]. This variability may be attributed to differing levels of endogenous hormones in different plant tissues. In moss rose (Portulaca grandiflora), the regeneration efficiency and optimal hormone concentration ratios differ among various explants, such as leaf discs and stem segments[47].
The physiological age of the explants is also one of the factors that cannot be ignored. For example, in the chrysanthemum cultivar 'Shinma', tissue-cultured plantlets with a physiological age of approximately six weeks demonstrated a higher regeneration efficiency than other age groups.[48] Generally, younger tissues present higher regenerative capacities than mature tissues[49]. In blackberry (Rubus idaeus), the bud regeneration significantly declined with increasing explant age; 7-day-old leaves regenerated more effectively than both 14-day and 28-day-old leaves[50]. In lily bulb scales, the younger cells at the base showed greater regenerative potential than those at the tip[51]. Nonetheless, the optimal physiological age for explants varies. For instance, 14-day-old leaves in Centratherum punctatum resulted in the highest shoot induction compared to leaves of other physiological ages[52], and 20-day-old leaf explants in Pogostemon quadrifolius exhibited the highest frequency of direct shoot induction[53]. A study on Populus deltoides revealed that the fifth leaf, aged 14−16 d, exhibited the highest regeneration potential compared to younger and older leaves[54]. Such internal factors contribute to the challenges encountered in studying the regeneration of ornamental plants.
Effects of environmental factors on ornamental plants regeneration
-
The regeneration of ornamental plants is influenced by various environmental factors[55]. The composition of the culture medium, including the concentration and types of inorganic salts, vitamins, and saccharides play a crucial role and varies according to the specific needs of each ornamental plant, thereby affecting their regenerative abilities[56].
After the invention of the MS medium by Murashige and Skoog in 1962, it found widespread application in the tissue culture of ornamental plants, including Sedum plumbizincicol, pyreturum, and moss rose[8,17,47]. Subsequently, in 1981, Lloyd and McCown improved the MS medium through the culture of Kalmia latifolia shoot tips, leading to the development of the WPM medium. Unlike the MS medium, the WPM medium substituted KNO3 with potassium sulfate decreased NH4NO3 content to one-fourth of that in the MS medium, and primarily delivered nitrogen salts in the form of calcium nitrate, rendering it more suitable for woody ornamental plants such as peony (Paeonia ostii) and eucalyptus (Eucalyptus grandis)[57,58]. Other media, like the B5 medium which boasts a higher vitamin content than the MS medium, are conducive to the regeneration of okra (Abelmoschus esculentus) and pepper (Capsicum annuum)[59,60]. In a research study focusing on aquatic ornamental plants, it was found that substituting MS medium with palm oil mill effluent (POME) not only decreases costs but also enhances the regeneration efficiency of Hemianthus callitrichoides more effectively. One contributing factor to this phenomenon is the threefold higher concentration of potassium particles found in POME than in MS medium[61]. Overall, various ornamental plants necessitate distinct optimal culture media; however, certain ornamental species remain incapable of regeneration in the current culture media.
Microbial contamination represents a substantial obstacle in plant regeneration, directly impacting the survival and success of genetic transformation in explants. Traditional methods involve sterilizing the growth medium and culture containers via high-temperature autoclaving. Nonetheless, this approach may result in the deterioration of crucial nutrients like amino acids and vitamins, along with the generation of harmful substances. Chemical disinfection, employing agents such as sodium hypochlorite, mercury, alcohol, or hydrogen peroxide, provides an alternative but risks harming the explants, thereby diminishing their regeneration capacity. In this context, the role of nanomaterials, particularly metal nanoparticles, does not inhibit plant growth has garnered scientific attention. For instance, optimal control of contamination was achieved through a 15 to 20-min treatment with 250 ppm AgNP on 4-week-old ex vitro chrysanthemum 'Jimba' leaves[62]. However, a study confirms that they can decrease the regeneration efficiency and impede the growth of Aldrovanda vesiculosa while Nano-silver particles can reduce the contamination rate in the tissue culture[63].
Environmental conditions in the culture room, specifically light and temperature, substantially impact plant regeneration. Appropriate photoperiods are usually specific to the species or genotype of various ornamental plants. For example, Narcissus tazetta exhibited greater success in vitro bulb formation under a 16/8 h light/dark photoperiod compared to complete darkness[64]. In contrast, for pyrethrum , a period of cultivation in darkness was more conducive to explants regeneration[24].
Light quality is another critical factor. A combination of higher red and lower blue LED light is suitable for plantlet regeneration in species like Phalaenopsis, rose (Rosa kordesii), chrysanthemum 'Ellen', gerbera (Gerbera jamesonii), Heuchera × hybrida, heliconia (Heliconia metallica), Ficus benjamina, and Lamprocapnos spectabilis[2,65]. However, in Vanilla planifolia, blue light (460 nm) was found to enhance shoot growth and chlorophyll synthesis[66]. The highest induction of Phalaenopsis pulcherrima protocorm-like bodies occurred under red and blue LED conditions[67]. Besides, low doses (60 s) of He-Ne laser irradiation exerts a positive effect on in vitro Vanilla planifolia regeneration and acclimation[68].
Temperature also plays a crucial role in plant regeneration[69]. For example, the optimal growth and survival rate of regenerated Fragaria × ananassa, plants were observed in a growth chamber maintained at 25 ± 2 °C[70]. This temperature range is generally suitable for most ornamental plants. However, there is also a studyc confirming that chrysanthemums exhibit enhanced budding ability in the ovary and ovule wall after callus tissue formation under both low (4°C) and high (32°C) temperature conditions[44].
Effect of plant growth regulators on plant regeneration
-
PGRs are pivotal in the in vitro regeneration of plants, profoundly influencing growth and developmental processes. Research on the model plant Arabidopsis has illustrated that plant growth and development are regulated by the synergistic interactions of various PGRs and endogenous hormones, including auxins (AUXs), cytokinins (CTKs), gibberellins (GA), abscisic acid (ABA), ethylene (ETH), and brassinosteroids (BR). These hormones, in concert with regenerative transcription factors, play critical roles in meristem maintenance, organogenesis, and modulating the expression of genes associated with regeneration[71]. Exogenous cytokinins prominently contribute to the induction of regenerative tissues through the indirect regeneration pathway in most ornamental plants, while exogenous auxins primarily drive the somatic embryo pathway. The combined presence of auxins and elevated levels of cytokinins can stimulate direct organogenesis[26,39,72]. In the context of exogenous plant growth regulators (PGRs) during organogenesis, 2,4-D is frequently omitted or utilized in significantly lower quantities compared to auxin. However, during the induction phase of somatic embryogenesis, 2,4-D serves as the primary PGRs, with concentrations exceeding other PGRs. Research has demonstrated that the using of 2,4-D can redirect explants engaged in organogenesis toward regeneration through somatic embryogenesis[73]. Consequently, high levels of 2,4-D are seldom employed in PGR applications during organogenesis, while in the somatic embryogenesis process, 2,4-D plays a prevalent and crucial role. Besides its growth regulatory function, 2,4-D also triggers somatic embryo formation by modulating plant signal transduction pathways[74]. In organogenesis, 2,4-D is typically excluded, and when occasionally used in minor quantities, it can often be substituted with NAA or alternative growth regulators.
In ornamental plant regeneration, Auxin and CTK are predominantly used in regeneration media due to their fundamental regulatory roles[34]. Other PGRs, such as GA, ABA, ETH, and BR, also play crucial roles in the somatic embryogenesis of specific ornamental species. For instance, exogenous GA has been found to stimulate the maturation of Catharanthus roseus, thereby enhancing somatic embryogenesis efficiency[75]. ABA has been shown to facilitate the maturation and regeneration of torpedo embryos in tulips[76], while ETH has been observed to increase the production of somatic embryos in stem-derived leaves of Doritaenopsis hybrid[77]. The addition of 24-epibrassinolide has been beneficial in generating rounded bulbs in orchid explants, enhancing their reproductive efficiency[78].
Besides, specific concentrations of Methyl Jasmonate (MeJA), particularly between 50–100 μM, have yielded promising results in increasing the fresh and dry weight of Phalaenopsis pulcherrima protocorm-like bodies (PLBs), as well as positively affecting the induction and number of PLBs per explant. However, it's imperative to note that higher concentrations of MeJA may inhibit plant growth and decrease PLB induction, highlighting the importance of precise dosage control[79]. Moreover, the incorporation of melatonin in regeneration media has been found to up-regulate genes like LoPCNA and LoCYCB2-1, while inhibiting the expression of LoKRP3 and LoTCP4 during the early stages of lily bulb development, thereby promoting bulb growth[80]. While PGRs play a crucial role in plant regeneration, certain ornamental species cannot regeneration despite efforts to adjust the PGR concentration and type.
-
The in vitro regeneration of ornamental plants is crucial for the study of ornamental plants. Nonetheless, there are still certain limiting factors that need to be addressed. These limitations primarily include somaclonal variation, difficulties in inducing shoot formation in callus tissues, substantial browning, and inability to produce adventitious roots, among others. Due to these constraints, some ornamental plants lack the potential for commercial production[81].
Somaclonal variation can occur in all types of tissues and result in a series of adverse effects on the populations formed by tissue culture[81]. For instance, in the cut chrysanthemum 'Arjuna', somatic lineages from leaf in vitro propagation showed significant differences in several indicators such as flower size, flower weight, leaf weight, and plant size compared to early measurements, and color changes in flowers and leaves occurred[82]. Furthermore, in the agrobacterium-mediated pyrethrum genetic transformation (clone No. 39), the pyrethrum material used in this method was a specific radiation-induced mutant, which subsequently lacked regenerative capabilities due to somatic embryogenesis mutation, making it irreproducible[17]. The causes of mutations could be multifaceted, such as changes in DNA methylation status, chromosomal ploidy differences due to cell proliferation, MicroRNA misregulation, and more[83]. Currently, in some ornamental plants, resolution methods mainly include visual inspection, using molecular markers, conducting cytogenetic investigations through Mass/Flow cytometry, and measuring changes in key genes related to methylation/acetylation[84]. Additionally, adjusting hormone types in culture media can potentially serve as a solution. In a study on Doritis pulcherrima regeneration, the addition of extra 2,4-D to somatic mutated plants significantly promoted regeneration[85]. In general, for some important ornamental plants, it is essential to detect the genetic structure and stability of regenerating plants and screen for somaclonal variation during tissue culture.
Plant regeneration is a highly complex process and some ornamental plants exhibit high regeneration capabilities controlled by multiple genetic factors. However, some ornamental plants cannot induce shoot formation after callus induction during the regeneration process, for example, Dendrocalamus brandisii and peony[57,86]. It has been confirmed in the model plant Arabidopsis that TRITHORAX-RELATED 2 (ATXR2) protein can regulate cytokinin signaling during cell division, prevent premature activation of WUSCHEL, ensure cell fate transition, and that the mutation atxr2-1 can enhance the regeneration ability of callus tissues[87]. In dwarf morning glory, ATXR2 shows higher expression levels in callus tissues[88]. In subsequent studies on ornamental plant regeneration, by mutating ATXR2 in difficult-to-regenerate materials, it may be possible to address the issue of callus formation without shoot induction. Additionally, during the induction process of callus tissue, the use of immature embryos instead of mature ones can partially replace traditional browning inhibitors, thus addressing the issue of callus tissue browning and enhancing the regeneration capacity of ornamental plants[89].
The market for ornamental plants depends on species diversity and the availability of high-quality breeding materials, and aseptic induction of adventitious roots (AR) plays a critical role in commercial propagation and maintenance. Generally, the rooting-inducing hormones commonly used in tissue culture environments are NAA and IBA, while CTKs negatively regulate adventitious root formation[90]. In Arabidopsis, the generation of reactive oxygen species (ROS) induced by wounds participates in AR induction by regulating auxin biosynthesis and transport[91]. In Azalea Microshoots, humic acid (HA) in explants can increase the level of ROS, thereby promoting AR production[92]. In conclusion, enhancing endogenous auxin biosynthesis and combining it with the application of plant growth regulators (PGRs) may represent a potential direction for addressing the issue of rooting difficulties in ornamental plant tissue culture.
-
Plant regeneration is a prerequisite for the establishment of genetic transformation systems. Traditionally, transgenic plants are equipped with resistance genes, enabling them to thrive in a culture medium containing specific screening substances, an ability lacking in non-transgenic plants. These substances, including kanamycin, hygromycin, phosphinothricin, and glyphosate, are known to inhibit plant protein synthesis. However, the studies involving numerous monocotyledonous ornamental plants have indicated low screening efficiencies with these traditional agents[93]. Exposure of untransformed cells to these substances typically result in browning and cell death, releasing phenols that substantially impede the regeneration of nearby transgenic cells. For example, research in alfalfa highlighted a potential 'escape' phenomenon in non-genetically modified plants, particularly when explants were not in direct contact with the screening medium[94].
In response to these challenges, innovative screening methods for transgenic ornamental plants have been developed. One notable strategy involves integrating genes that enhance regeneration directly into the Ti plasmid, using regenerative genes as screening markers. A case in point is the integration of an overexpressed GRF–GIF chimeric protein sequence with the CRISPR/Cas9 system into a T-DNA region for wheat (Triticum aestivum) callus transformation. This method generated transgenic plants without the need for cytokinin and selectable markers[95] (Fig. 2a). Similarly, the CRISPR-Combo system's editing of the target gene and activation of the rice regeneration-related gene OsBBM facilitated successful plant regeneration under PGR-free conditions, while untransformed calli failed to regenerate[96]. In the genetic transformation of snapdragon (Antirrhinum majus), integrating the PLT5 gene into the carrier T-DNA promoted the development of adventitious buds at stem wound sites that exhibited stable transgenic traits transferable to subsequent generations. In contrast, the control group lacking integrated PLT5 genes showed no signs of organogenesis[97] (Fig. 2b).
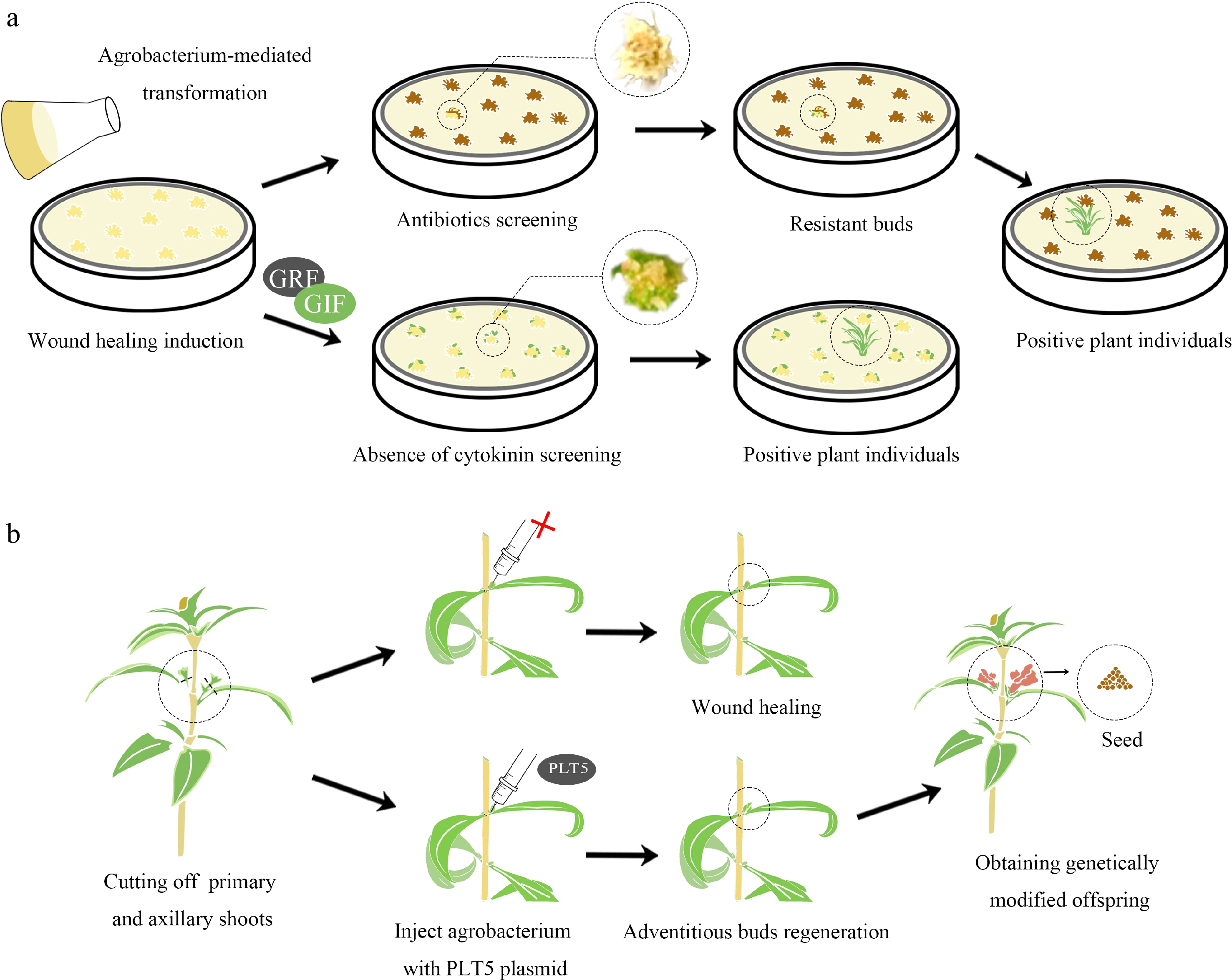
Figure 2.
Screening genetically modified plants by enhancing their regeneration ability. (a) Traditionally, the process of transformational screening involves the introduction of Agrobacterium into plant explants, which carries a resistance gene in the T-DNA region of its plasmid. These explants are then screened in specific antibiotics to obtain resistant shoots. When explants are infected with GRF-GIF chimeric protein, they are placed in a cytokinin-free medium for regeneration. Shoots produced without the need for cytokinins could potentially be positive and selected in an antibiotic-free manner. (b) Snapdragons were injected with plasmids containing or lacking the PLT5 gene. After injection with agrobacterium carrying the PLT5 gene, small buds formed at the wound sites of the goldfish plants, which could give rise to positive seeds. In contrast, wounds injected with Agrobacterium lacking the PLT5 gene healed without bud formation.
In brief, these advanced approaches in screening genetically modified plants, which focus on promoting the complete regeneration of modified plants rather than inhibiting the regeneration of unmodified plants, are not only time-efficient but also highly effective[96]. This methodology proves particularly beneficial for plant species with inherently low regeneration capacities.
Application of genes related to regeneration
-
In the late 20th century, there was a witnessed direction shift in plant-regeneration research, with the emergence of molecular genetics and transcriptomics leading to a focus on regeneration-related genes and their signaling pathways. This shift underscored the importance of the orderly regulation of these genes during plant regeneration. The capacity for direct regeneration in explants is determined by the presence of renewable cells, which is governed by the expression levels of specific regenerative genes. Altering the expression of these genes in non-renewable cells can convert them into renewable cells. Subsequent in-depth research has identified key genes instrumental in model plant regeneration, including WUSCHEL (WUS), WUSCHEL HOMEOBOX (WOX), SHOOT MERISTEMLESS (STM), BABY BOOM (BBM), and PLETHORA (PLT)[98] (Fig. 3).
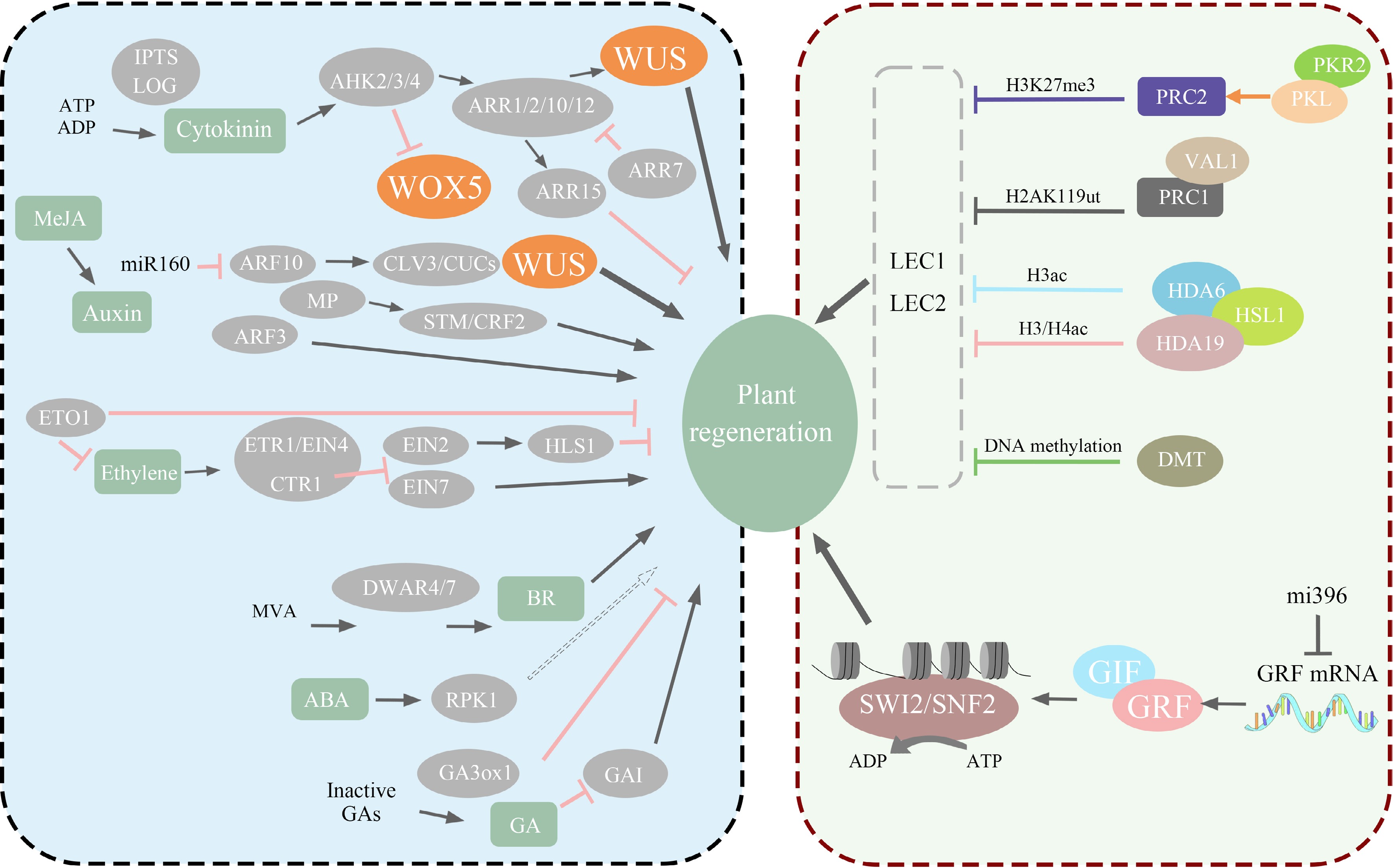
Figure 3.
The left half of the diagram illustrates how various genes related to regeneration directly or indirectly regulate plant regeneration through different pathways, such as WUS and WOX. The right half of the diagram portrays the impact of epigenetics on plant regeneration, including histone modifications and nucleosome structure remodeling.
While these regeneration-related genes are crucial for plant regeneration, it is important to note that their functions exhibit variations and are not entirely identical. WUS is essential for maintaining the stem cell niche in the shoot apical meristem. It facilitates the differentiation of lateral primordia and supports the totipotency of plant cells[99]. WOX plays a vital role in regulating the production and maturation of meristem cells, thereby impacting plant growth and development[100]. Distinct from WUS, STM functions in the specification of the apical meristem boundary region and in maintaining meristem activity by inhibiting processes such as leaf and cell differentiation[101]. Additionally, BBM influences the expression of auxin synthesis genes and promotes somatic embryogenesis through enhanced auxin response and biosynthesis[102]. Similar to the BBM function, PLT directly regulates PIN-mediated auxin transport and synthesis, establishing the hormonal polarity necessary for regeneration[103] (Fig. 3).
These genes have been effectively utilized to improve industrial crops regeneration efficiencies. For instance, the application of TaWOX5 has improved wheat immature embryo genetic transformation and regeneration[104]. MtWOX9-1 in Medicago truncatula stimulates somatic embryogenesis[105], and MdWOX4-2 in apple (Malus domestica) has been shown to improve leaf regeneration efficiencies and form adventitious shoots[106]. Co-expression of WUS and BBM has facilitated the redifferentiation of mature rice and maize leaves[107], challenging the traditional belief that mature leaves are incapable of initiating callus formation or regenerating into complete plants.
Compared to crops and model plants, small amounts of regeneration genes are also beginning to be used in the regeneration of ornamental plants. PLT5, for example, has been reported to aid in callus formation and regeneration at damaged stem sites in snapdragon[97]. The overexpression of PpWOX2 in Pinus pinaster has altered embryogenesis-associated gene expression, promoting somatic embryo formation[108]. Similarly, BBM overexpression significantly improved the efficiency of somatic embryogenesis in switchgrass (Panicum virgatum)[107]. Furthermore, atypical genes that enhance regeneration efficiency have been identified, such as LiMYB75 in crape myrtus (Lagerstroemia indica), which stimulates STM expression and indirectly influences regeneration[109]. With the emergence of single-cell sequencing and spatial transcriptomics technologies, both the identification of regeneration-related genes in various ornamental plants and the analysis of the reasons for poor regeneration by revealing spatial relationships between different cells are facilitated[110]. This progress holds promise for the regeneration of ornamental plants that have proven challenging to regenerate.
Promotion of plant regeneration through epigenetic modification
-
Epigenetic modifications in plants, alterations in DNA methylation levels DNA, histone modification, and chromatin remodeling, have become increasingly significant in understanding plant regeneration processes[111]. The regeneration process of explants are influenced by changes in their epigenetic status[112]. The in vitro plant regeneration process is known to induce alterations in epigenetic states, influenced by the expression of nuclear factor Y (NF-Y) and transcription factors LEAFY COTYLEDON1 (LEC1) and LEAFY COTYLEDON2 (LEC2), which are integral to developmental regulatory processes[113], such epigenetic changes play a significant role in the process of plant regeneration[7] (Fig. 3).
Alterations to DNA methylation levels, among various epigenetic modifications, are crucial in plant regeneration. Research on model plants suggests that apical meristem stem cells may serve as transposon loci, and DNA demethylation can activate transposons[114]. The expression of the regeneration-related key transcription factor WUS, essential in stem regeneration, is regulated by 5mC methylation during organ formation[115]. Additionally, histone modifications are critically involved in regeneration. Tissue culture induces the reprogramming of histone H3K27me3 modifications, crucial for callus formation[116]. Explant wounding is regarded as a crucial aspect of regeneration[117], wound-induced regeneration involves heightened histone H3 acetylation (H3Ac), triggering gene deactivation at the wound site for a rapid response[118]. In Arabidopsis, jasmonic acid (JA) regulates histone methylation levels, enhances auxin expression, and facilitates root development.[119] The expression of WOX11 and WOX12, important in regeneration, is regulated by H3K27me3, with their expression upregulated following the removal of this histone modification[120]. Furthermore, H3K4me2 demethylation aids the callus in responding to exogenous signals and initiating stem differentiation, enhancing regeneration[121]. Recently, there has been increasing attention to the link between chromatin remodeling and regeneration. The SWI/SNF complex utilizes ATP hydrolysis to modify the positioning of nucleosomes, thereby reshaping chromatin[122], a process that is vital for maintaining the apical meristem[123] (Fig. 3).
Chimeric transcription factors
-
The development of chimeric transcription factors created through protein fusion techniques, significantly enhances the effectiveness of regeneration-related transcription factors. VP16, stabilizes the transcription initiation complex and, when localized to the promoter of a target gene, can significantly up-regulate gene expression[124]. The resultant VP16–WUS chimeric protein in Arabidopsis showed an enhanced stem-tip meristem phenotype, exhibiting a stronger effect compared to the overexpression of WUS alone[125].
Recently, the GRF-GIF chimeric protein has presented another innovative approach. GRF binds to specific regeneration-related target DNA sequences, while GIF can recruit SWI/SNF chromatin remodeling complexes. The GRF–GIF chimeric protein can remodel chromatin at target DNA sites, effectively displacing nucleosomes from the promoter region of regeneration-related genes thereby increasing their expression[126].
Significantly, despite the observed strong species and genotype-dependent nature of plant regeneration[34], GRF–GIF chimeric protein has exhibited high compatibility across diverse species and regeneration pathways. Crucially, the ectopic expression of these fusion proteins has not been found to negatively impact plant development[127]. For instance, the GRF4–GIF1 chimeric protein in wheat has been demonstrated to enhance regeneration efficiency in both somatic embryogenic pathways in monocotyledonous plants and organogenic pathways in dicotyledonous plants, such as citrus (Citrus reticulata)[95].
GRF–GIF chimeric protein has also found applications in the regeneration of ornamental plants. A notable example is the use of PeGRF6–PeGIF1 in Phalaenopsis equestris, which regulates the proliferation of leaf cells[128]. In the investigation of the succulent plant Sedum plumbizincicola, it was observed that the overexpression of SpGRF4-SpGIF1 led to vitrification under tissue culture conditions, impeding shoot development. By employing the Cre/loxP self-excision system, not only was the regeneration and transformation of S. plumbizincicola facilitated, but the issue of explants vitrification was also circumvented[8]. Hence, the successful application of chimeric transcription factors in model plants and crops provides a prospective theoretical framework for resolving regeneration challenges in ornamental plants[127].
-
The CRISPR/dCas9 system represents a significant advancement in plant science, particularly in the activation of regeneration-related genes and the modification of epigenetic states to improve plant regenerative potential. Utilizing CRISPR/dCas9 system, such as targeting VP64 near the promoter of regeneration-related genes, can enhance gene expression and consequently improve plant regenerative capacity. An example of this is the CRISPR-Combo system, derived from CRISPR/dCas9 system, which activates the expression of the OsBBM gene in rice, facilitating somatic embryogenesis without the need for PGRs[96,129].
Targeted epigenetic modification tools based on CRISPR/dCas9 system have been successfully applied in Arabidopsis for DNA methylation changes[10,130]. However, their use in altering epigenetic modifications related to ornamental plant regeneration has yet to be reported. In ornamental plants, such as in the demethylation of the CmMYB6 promoter in chrysanthemums using the dCas9-TET1cd system, this approach changed the flower color from yellow to pink[11]. In the future, the CRISPR/dCas9 system holds promise in addressing the challenge of regeneration difficulties in certain ornamental plants.
Hormone-induced changes of chromatin accessibility in somatic embryos
-
Chromatin accessibility, influenced by factors like nucleosome occupancy and tissue dynamics, plays a crucial role in gene expression levels and subsequent cellular functions[131]. In Arabidopsis, auxin has been shown to modify chromatin accessibility, thus promoting cell totipotency and facilitating regeneration[132]. Follow-up research in Arabidopsis revealed that an environment with a high auxin/cytokinin ratio can manage the accessibility of chromatin, thus facilitating shoot production[133]. In wheat, auxin-induced gene expression changes through alterations in chromatin accessibility and histone methylation statuses (H3K27me3 and H3K4me3) have been observed. Techniques like RNA sequencing and transposase-accessible chromatin sequencing have identified potential genes, TaDOF5.6 and TaDOF3.4, as drivers of wheat regeneration and transformation[12].
Chromatin accessibility dynamics have also been observed in ornamental plants, such as Populus spp. under various stress conditions[134]. Moreover, the accessibility of hemiparasite Santalum album chromatin is modulated by bioactive compounds like melatonin in a study[135]. While studies directly linking chromatin accessibility to regeneration in ornamental plants are currently lacking, this area is poised to become a new research frontier in enhancing regeneration capabilities in these species.
Regeneration and transformation methods independent of tissue culture
-
The utilization of regeneration-related genes and gene editing techniques has led to significant advancements in plant regeneration and genetic improvement. Nonetheless, the genetic transformation of most plants through these methods is hindered by genotype limitations[13]. Moreover, the tissue culture procedure is characterized by its time-consuming and intricate nature. Presently, alternative transformation approaches that do not rely on tissue culture, like the floral dipping transformation protocol employed in Arabidopsis, are available[136]. Nevertheless, this methodology might not be suitable for the majority of ornamental plants.
The cut-dip-budding system is utilized on specific plant root systems with the potential to produce adventitious buds. These particular plants are known as root-suckering plants. Under controlled conditions, plant regeneration can be induced by inoculating them with Agrobacterium rhizogenes, which stimulates the growth of adventitious roots. After the emergence of adventitious roots from the parent plant, they develop into shoot primordia, which can further propagate into independent plants. This method has been successfully utilized in the genetic modification and gene editing of various medicinal plants and crops[13,137]. Moreover, it is well-suited for certain succulent ornamental plants like Kalanchoe blossfeldiana, Crassula arborescens, and Sansevieria trifasciata, which all exhibit the shared trait of being able to generate new shoots from their leaves[138].
The Regenerative Activity-dependent in Planta Injection Delivery (Rapid) system is another method that leverages the active regenerative capability of plants. By directly injecting agrobacteria into plant explants, transformation can be achieved without the reliance on tissue culture[139]. This system has been successfully applied to a variety of plants including sweet potato (Ipomoea batatas) and potato (Solanum tuberosum). In future studies, it may be considered to use the above method for the genetic transformation of plants with strong regenerative abilities in their stems or leaves, chrysanthemums and begonia (Begonia grandis). Even plant varieties that pose challenges in regeneration and transformation through traditional tissue culture methods may benefit from this approach for future transgenic transformation purposes.
Recent reports confirm that directly injecting WUS2 combine with STM into tobacco (Nicotiana benthamiana) can efficiently promote genetic transformation and gene editing in tobacco without the need for tissue culture, a method known as Fast-TrACC[140]. With ongoing validation of regeneration-related genes in ornamental plants, combining regeneration genes with transformation independent of tissue culture holds the potential to enable the regeneration and transformation of ornamental plants that are difficult to regenerate and laborious to culture.
-
High-quality ornamental plants can be propagated in vitro to generate sufficient raw materials, ensuring the large-scale production of ornamental plants. By utilizing appropriate culture media, optimizing the use of RGRs, and meticulously controlling environmental conditions in vitro, the regeneration of select ornamental plants can be facilitated. Nonetheless, the vast array of ornamental plant species, coupled with the intricate plant regeneration, presents challenges in formulating generalized methodologies applicable across all ornamental plant types, given the complexities observed in many studies. Regenerating many crucial ornamental plants remains a formidable task.
During the regeneration process of ornamental plants, challenges often arise, such as somaclonal variation, difficulty in shoot formation in callus tissue, extensive browning, and failure to develop adventitious roots. Due to the complexity of regeneration, there is no one-size-fits-all solution applicable to all ornamental plants. This work presents potential methods to address the issues encountered and the solutions employed during the regeneration process of ornamental plants. These methodologies encompass alterations in screening agents, the integration of regeneration-related genes, the application of epigenetic modification, and the utilization of chimeric proteins to advance ornamental plant regeneration. Furthermore, this paper suggests activating regeneration genes through the CRISPR/dCas9 activation system, modifying chromatin accessibility, and combining regeneration-related genes with non-tissue culture methods to bolster the regeneration and genetic refinement of challenging ornamental plant species. These innovative strategies offer new pathways for conserving and expanding rare cultivars, as well as enriching the diversity of new varieties.
-
The authors confirm contribution to the paper as follows: study conception and design: Wang C, Zeng T, Zhu L, Zhou L; data collection: Zhu L, Zhou L; analysis and interpretation of results: Zhu L, Zhou L, Li J; draft manuscript preparation: Zhu L, Zhou L, Zeng T, Wang C; feedback on the analysis and manuscript: Chen Z, Wang M, Li B, Xu S, Luo J, Zeng T, Wang C. All authors reviewed the results and approved the final version of the manuscript.
-
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
This work was supported by the Innovation Demonstration Base of Hubei Macheng Chrysanthemum Industry Science and Technology (Grant No. 305-707122125); National Natural Science Foundation of China (32160718); Natural Science Foundation of Guizhou Province [ZK [2022] 301].
-
The authors declare that they have no conflict of interest.
-
# Authors contributed equally: Liyong Zhu, Li Zhou
- Copyright: © 2024 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Zhu L, Zhou L, Li J, Chen Z, Wang M, et al. 2024. Regeneration of ornamental plants: current status and prospects. Ornamental Plant Research 4: e022 doi: 10.48130/opr-0024-0022
Regeneration of ornamental plants: current status and prospects
- Received: 13 March 2024
- Revised: 04 July 2024
- Accepted: 11 July 2024
- Published online: 07 August 2024
Abstract: For horticulture to meet the needs of global markets, the reproduction and breeding of ornamental plants is allimportant. Nevertheless, certain ornamental plants exhibit a lower capacity for explant regeneration when compared to model plants. These challenges hinder the rapid propagation, virus-free breeding, and molecular breeding of ornamental plants. This paper examines both traditional and emerging plant regeneration technologies and discusses the difficulties ornamental plants encounter during the regeneration process. It also provides an outlook on the applications of emerging technologies in ornamental plant regeneration. This study will provide insights into the industrialization and practical application of molecular tools in ornamental plant breeding.
-
Key words:
- Ornamental plant /
- Current research /
- Current bottleneck /
- Regeneration techniques












