-
Ginkgo biloba is one of the oldest extant seed plants in the world, and it has high ecological and economic significance. G. biloba is widely used as a fruit tree, ornamental and medicinal tree species with a long life span (e.g., in China, Japan, Korea, and France)[1]. Because the leaves and seeds are rich in flavonoids and terpenoids, Ginkgo biloba extract (GbE) is used in medicine to treat cardiovascular and cerebrovascular diseases and cancer, and it is also used as a food supplement and nutraceutical. The flavonoid content is a key factor affecting the quality of GbE[2,3]. In G. biloba, flavonoid accumulation is affected by environmental factors such as light, temperature, and water, as well as biotic or abiotic stresses such as drought, cold, and ultraviolet light[4,5]. Furthermore, many studies have demonstrated that age can affect the flavonoid content in the leaves of G. biloba[6]. Due to the publication of the G. biloba genome and continuous improvement in both molecular biology and sequencing technologies, research on the molecular mechanism of flavonoid synthesis has greatly advanced. Our paper focuses on the regulation mechanisms affecting flavonoid biosynthesis in G. biloba, in particular, the impact of structural genes, transcription factors, and non-coding RNAs. To further explore the characteristics and regulation mechanisms of flavonoids, we also analyzed their functions, types, synthetic pathways, and external influencing factors. This review aims to summarize research on the regulatory network of flavonoids and promote research on the synthesis and accumulation of flavonoids in G. biloba.
-
Flavonoids include flavonols, flavanols, flavones, flavanones, and isoflavones, which are distinguished based on the structural characteristics of the C-ring[7]. In G. biloba, the flavonoids mainly consist of the glycosides biflavone and monoflavone. Monoflavones are the most important components, and are made up primarily of kaempferol, quercetin, and isorhamnetin[8]. The biflavones in G. biloba include ginkgetin, isoginkgetin, sciadopitysin, and bilobetin. In addition, six types of catechins have been isolated from G. biloba: catechin, gallic catechin, epicatechin, epigallocatechin, 4,8' gallic catechin gallic catechin, and 4,8' catechin gallic catechin[9].
As the important secondary metabolites, flavonoids can resist biotic and abiotic stresses[10]. Previous studies had found that flavonoids had protective functions by absorbing ultraviolet (UV)-B radiation and clearing reactive oxygen species[11,12]. As evidence, the flavonol content of leaves was highest at the highest total solar radiation intensity, suggesting that high levels of UV radiation enhance the flavonol accumulation[13]. Researchers also found that the flavonoids in G. biloba can remove free radicals to resist salt stress and low temperatures[5,14]. In terms of drought, the flavonoid content of G. biloba increased with decreasing relative soil water content under an early dry stress of < 35% relative soil water content, indicating the function of flavonoids in drought resistance[15]. The flavonoids in G. biloba can also act as a defense against insects. Compared with mechanical damage, herbivore wounding increased the glycosylated flavonoid content by nearly two times. The expressions of key genes regulating flavonoid synthesis, such as phenylalanine ammonia-lyase (PAL), flavanone 3-hydroxylase (F3H) and anthocyanidin reductase (ANR), were significantly up-regulated after herbivore wounding[16]. More importantly, several genes related to defense mechanisms were identified in the G. biloba genome, 29 of which are also involved in the core pathway of flavonoid synthesis[17]. Due to the fact that most of the trees died from pests and diseases or from environmental stresses such as drought, but the accumulated protective metabolites including flavonoids in trees may enhance their resistance to the stresses, it is therefore speculated that the accumulation of flavonoids may be one of the reasons for G. biloba longevity[18].
Flavonoids of G. biloba have high medical value and are of great importance to human health[19,20]. An increasing number of studies indicate that the anti-cardiovascular activity of GbE is mainly determined by flavonoids[21,22]. The biflavonoids in G. biloba may be anticarcinogenic by inhibiting tissue kininogenin and autophagy[3,23]. Furthermore, ginkgetins can suppress cancer by blocking the cell cycle, inducing cell apoptosis and other signaling pathways, and protecting against influenza viruses by inhibiting sialidase activity[24,25]. The monoflavone of G. biloba has significant antibacterial properties against multiple Gram-positive and Gram-negative bacteria[26]. In particular, kaempferol in G. biloba can reduce serotonin breakdown and reduce apoptosis through monoamine oxidase[2,27]. Moreover, isorhamnetin and quercetin of G. biloba have important roles in anti-inflammatory and antioxidant processes[28]. Specifically, isorhamnetin can reduce cell apoptosis and DNA fragmentation, which is beneficial to the cardiovascular and cerebrovascular systems[29,30]. In view of these, flavonoids in G. biloba play a significant and unique role in resisting environmental stress, and have advantages in promoting human health (Fig. 1).
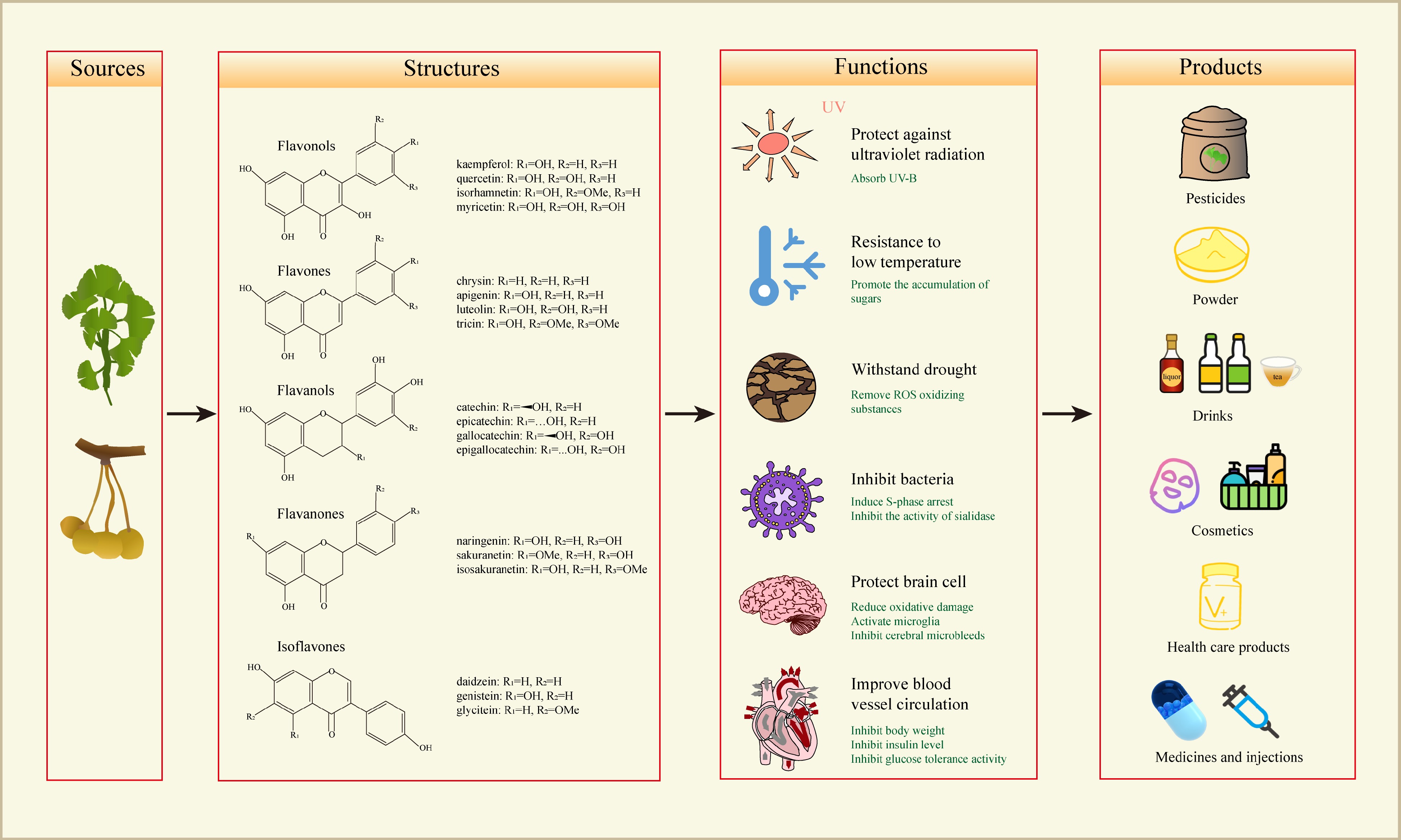
Figure 1.
The structures, functions and products of the flavonoids in G. biloba. Flavonoids in G. biloba are mainly divided into these categories, including flavones, flavonols, flavanols, flavanones and isoflavones. The flavonoids have the functions of inhibiting bacteria, withstanding drought, resisting low temperatures and protecting against ultraviolet radiation, as well as human health benefits for protecting brain cells and improving blood vessel circulation. In production, G. biloba flavonoid can be made into pesticides, powder, drinks, cosmetics, health care products, medicines and injections.
-
There are significant differences in the accumulation of flavonoids in different G. biloba organs and developmental stages[31,32]. The flavonoid content in leaves is higher than that in branches and exotestal[33]. Matrix-assisted laser desorption/ionization (MALDI) mass spectrometry (MS) was used to study the distribution of flavonoids in specific organs of G. biloba, and the content of flavonoid glycosides was found to be higher around the vascular chain of leaves[34]. The spatial chemical localization of flavonoids in G. biloba showed that flavonoids, especially the cyclodimers of flavonoids, are mainly located in the leaf epidermis[35,36].
The total flavonoid content in G. biloba leaves differs by tree age. Through a comparative analysis of one- to seven-year-old G. biloba trees, it was found that the flavonoid content of G. biloba decreases with increasing tree age[6]. Another study that used transcriptomics and metabolomics to analyze the leaves of one-, four-, and seven-year-old G. biloba trees also showed that 82% of flavonoid metabolites decreased with age, indicating that age is negatively related to flavonoid content[6]. Due to the high content of flavonoid in the leaves of young trees, the leaves of one- to five-year-old G. biloba trees are usually used as raw materials for medicine and food supplement. However, rejuvenating can promote the accumulation of flavonoids in G. biloba. Investigating a 544-year-old G. biloba tree with vigorous resprouters revealed that the thickness, fresh weight, and number of leaves, as well as the contents of kaempferol and isorhamnetin increased significantly in resprouter leaves. Further transcriptome analysis showed that the expression levels of genes related to flavonoid synthesis, including PAL and flavonol synthase (FLS), increased significantly in resprouter leaves[37]. These results suggest that even in old G. biloba trees, rejuvenation can also effectively activate the accumulation of flavonoids. Similarly, an additional study also found that after truncation treatments, the leaves of renewed G. biloba leaves were larger and thicker than control, the accumulation of flavonoids increased significantly and that the expressions of genes related to flavonoid synthesis, including chalcone synthase (CHS), FLS, flavanone 3'-hydroxylase (F3'H), and dihydroflavonol 4-reductase (DFR), were upregulated[38]. Further studies showed that the endogenous hormone levels changed after rejuvenation. By analyzing cis-acting elements of promoters of genes related to flavonoid synthesis, it was also found that these genes may be regulated by hormonal responses. For example, auxin (IAA) can promote the expression of GbFLS7, GbDFR11 and GbANS5, gibberellin (GA3) treatment also promotes the expression of GbPAL8 and Gb4CL5[39]. These results reveal that the flavonoid contents of G. biloba leaves can be increased effectively by rejuvenation through responding to hormones, which is an effective and feasible method to increase flavonoid contents.
-
Different environmental factors and stresses cause different physiological and metabolic responses in G. biloba[40]. Light, including light intensity, light quality, and photoperiod, are essential factors for plant growth and development, as well as the most important environmental factor that affects flavonoid accumulation. Light with different energies has different influences on the accumulation of flavonoids in G. biloba. Quercetin, kaempferol, and total flavonoid contents in G. biloba leaves under blue light were higher than those under mixed light, white light, and red light[41]. Interestingly, when treated with salicylic acid (SA), the synthesis and accumulation of total flavonoids in the leaves decreased in the dark while the flavonoid content increased under light conditions. In addition, red and far-red light are essential in SA-induced flavonoid accumulation, demonstrating the importance of light in flavonoid accumulation in G. biloba[42]. UV radiation is one of the main enhancers of flavonoid contents[12]. The sensitivity of G. biloba leaves to UV-B radiation varies at different developmental stages; young leaves can quickly establish a protective mechanism against harmful radiation, thus increasing their flavonoid contents[43]. Elevation also affects the flavonoid content. Several key genes involved in flavonoid biosynthesis, including DFR, leucoanthocyanidin reductase (LAR) and ANR, were significantly upregulated at higher elevation (with high UV radiation) in G. biloba[44]. According to the results, light is an important factor in determining the accumulation of flavonoids, especially blue light and moderate ultraviolet radiation can effectively increase the accumulation of flavonoids, especially, combined with spraying SA or JA can further increase the flavonoid content in G. biloba leaves.
The influence of temperature on flavonoid metabolism is complex. To investigate how different combinations of day and night temperatures influence the flavonoid contents in G. biloba, multiple combinations of day and night temperatures (5/10 °C, 25/20 °C, and 35/30 °C [day/night]) were used to treat G. biloba seedlings. PAL activity, the key enzyme in the flavonoid synthesis pathway, was enhanced at 15/10 °C and inhibited at 35/30 °C, indicating that the lower temperatures were more beneficial for the accumulation of flavonoids in G. biloba[4].
Water also plays a crucial role in the synthesis and accumulation of flavonoids. The chlorophyll concentration in G. biloba leaves decreased and the total flavonoid concentration increased under treatment with full root-zone and partial root-zone drought. Moreover, the total flavonoid content under partial root-zone drought treatment was significantly higher than that under full root-zone drought treatment[45,46]. Based on these results, reduced irrigation and the maintenance of local root drought can not only reduce rotten roots but also promote the accumulation of flavonoids, which is an effective method to improve the flavonoid accumulation in G. biloba.
Several previous studies found that G. biloba can reduce the damage caused by salt stress by enhancing its antioxidant capacities. At increasing NaCl concentrations (treatment with 100 mmol/L or 200 mmol/L of NaCl), the SOD, POD, CAT, and flavonoid contents increased in leaves[5]. These results indicate that G. biloba can reduce salt damage by accumulating flavonoids and other active substances following salt stress treatments. Because the expression levels of GbF3H3, GbDFR3, GbLAR1 and GbLAR6 were significantly increased after abscisic acid (ABA) treatments, and the expression levels of Gb4CL3, Gb4CL14, GbDFR4 and GbANR5 were also significantly increased after methyl jasmonate (MeJA) treatments[39]. It is also possible that the increase of flavonoid synthesis under stresses is regulated by ABA or JA, but the specific regulatory mechanism is still not clear.
-
With the application of the third-generation sequencing technology, G. biloba genome has been further sequenced and assembled. A total of 27,832 protein-coding genes were obtained[47]. Using specific-locus amplified fragment sequencing (SLAF-seq), a high-density genetic map of G. biloba was constructed[48]. Based on the publication of G. biloba genome, 13 structural gene families have been identified to participate in the synthesis of flavonoids, and a large number of clones have been obtained, including GbPAL (11 genes), GbC4H (six genes), Gb4CL (15 genes), GbCHS (14 genes), GbFLS (10 genes), GbDFR (12 genes), GbANS (13 genes), GbANR (seven genes), GbCHI (three genes), GbF3H (six genes), GbF3’H (four genes), GbF3’5’H (four genes) and GbLAR (six genes). The expansion of structural multiple genes may be the a significant reason for the promoting the flavonoid synthesis in G. biloba[39].
PAL encodes the first key enzyme in the phenylpropane metabolic pathway; it catalyzes the conversion of phenylalanine to cinnamic acid, the precursor of secondary metabolites[49, 50]. In G. biloba, GbPAL is expressed in all tissues, with the highest expression in leaves and stems but the lowest expression in roots[51]. UV-B and salt stress can induce the expression of GbPAL[52]. GbCHS encodes another key enzyme in the synthesis of flavonoids in G. biloba. The transcription level of GbCHS was shown to be related to genetic, hormone, and light conditions, which positively correlates with the flavonoid content of G. biloba leaves[53]. Subcellular localization experiments showed that GbCHS is localized in the cytoplasm, nucleus, and cell membrane[39]. Overexpressed GbCHS callus of G. biloba, significantly increased the total flavonoid content, confirming that GbCHS plays an important role in the biosynthesis of flavonoids in G. biloba[38].
GbF3H contains DIOX_N and 2OG-Fell_Oxy domains, encoding a core enzyme at the branching sites of flavanones. GbF3H expression is highest in leaves and absents in roots[42]. Differently, GbFLS is expressed in both roots, stems, leaves and seeds, and in vitro enzyme activity assay showed that recombinant GbFLS protein not only catalyzed the formation of dihydrokaempferol to kaempferol, but also promoted the conversion of kaempferol from naringenin[54]. However, the genetic transformation system of G. biloba has not been established, the function of key genes regulating flavonoid synthesis can only be verified by heterologous transformation or transformation of G. biloba callus at present. In the future, it is necessary to explore the homologous overexpression and virus-induced gene silencing (VIGS) transformation system of G. biloba for identification and functional study of these identified candidate genes.
-
The regulation of flavonoid metabolism is mainly based on the regulation of transcription. Transcription factors bind to structural gene promoters and directly regulate the expression of flavonoid synthase genes. MYB, bHLH, and WD40 are important and well-studied transcription factors in the flavonoid synthesis pathway[55]. The MYB family is one of the largest transcription factor families in plants and plays an important role in the plant development and secondary metabolites biosynthesis[56]. Several binding sites of MYB were identified to be exist on the promoter region of GbANS, indicating that MYB may bind directly its promoter to regulate the flavonoid biosynthesis in G. biloba[57]. Through the transformation of G. biloba callus, it was found that the total flavonoid content in G. biloba callus overexpressed with GbMYBFL was significantly higher than that in the control, suggesting that GbMYBFL could positively regulate the flavonoid biosynthesis[58], while GbMYBF2 and GbMYBR1 may inhibit flavonoid biosynthesis[59,60], suggesting that MYB has different regulatory effects on flavonoid biosynthesis in G. biloba. Although these GbMYBs may influence flavonoid biosynthesis and accumulation, the potential functions need more experimental evidence.
MYB functions in the regulation of flavonoid biosynthesis usually binding to bHLH and WD40 proteins to form MBW complexes[61]. bHLH transcription factors are a class of transcription factors containing the basic helix-loop-helix domain. They exist widely in plants and play a crucial role in plant growth, development, and signal transduction[62]. bHLH transcription factors regulate the flavonoid synthesis by interacting with MYB transcription factors to activate target genes involved in flavonoid synthesis[63]. The WD40 transcription factor, also known as WDR protein, is an ancient and structurally stable protein[64]. Based on a correlation analysis between the Fragments Per Kilobase of transcript per Million fragments mapped (FPKM) value of WD40 gene and the flavonoid content in different tissues of G. biloba, six GbWD40 genes that might be involved in flavonoid metabolism were identified[65]. A novel WD40 gene named GbLWD1-like has been cloned in Ginkgo biloba. The gene is mainly expressed in the leaves of G. biloba, followed by the roots. However, there was no significant difference in the content of flavonoids in the leaves of overexpressed transgenic lines[66]. It is speculated that it may not require the participation of WD40 in the regulation of flavonoid biosynthesis in G. biloba, but more evidence is needed to verify the possible functions of WD40 related to flavonoid accumulation.
Flavonoid synthesis in G. biloba is also regulated by the transcription factors NAC (N-acetylcysteine)[67], bZIP (basic region-leucine zipper)[68], and TCP (Teosinte branched1/Cycloidea/Proliferating cell factor)[69]. The NAC family of transcription factors is one of the largest families of plant-specific transcription factors, and plays an important role in plant growth and development as well as biotic and abiotic stresses. By analyzing the NAC family in G. biloba, it was found that GbNAC007 and GbNAC008 were significantly correlated with flavonoid content, suggesting that GbNAC007 and GbNAC008 were involved in flavonoid biosynthesis[70]. The bZIP gene family is one of the largest gene families in plants. It is involved in secondary metabolism, stress responses, and seed maturation. A total of 40 bZIP genes were identified in the G. biloba. Correlation and phylogenetic tree analyses indicate that GbbZIP08 and GbbZIP15 might be involved in the biosynthesis of flavonoids[71]. The TCP transcription factors play an important role in plant growth and development, hormone signaling, biological clock signaling, stress responses, and secondary metabolism regulation[72]. Thirteen TCP genes were identified in the genome of G. biloba and phylogenetic analysis showed that five of those belonged to the PCF subclade and the rest to the CIN subclade. Based on a correlation analysis between GbTCP expression and flavonoid content, GbTCP03, GbTCP04, and GbTCP07 might be involved in flavonoid biosynthesis[73]. An increasing number of different types of transcription factors have been discovered to be involved in the regulation of flavonoid metabolism in G. biloba, but whether these transcription factors act directly on structural genes or interact with other transcription factors to affect the regulation of flavonoid synthesis is unclear. Yeast one-hybrid (Y1H), electrophoretic mobility shift assay (EMSA), and the dual luciferase assays can be used to further explore the regulation mechanisms of transcription factors on genes related to G. biloba flavonoids. In summary, the current research on the regulation of flavonoid synthesis in G. biloba is still limited to the identification of key genes through some omics analysis, but the function of key genes and their regulatory mechanisms are still unclear. However, the study on the regulation mechanism of flavonoids will be of great significant to the biological breeding of high flavonoids in G. biloba.
-
Non-coding RNAs (ncRNAs) are a class of RNA molecules that do not encode proteins and have catalytic activity. NcRNAs can be mainly divided into microRNAs (miRNAs) and long-stranded ncRNAs (lncRNAs). MiRNAs are important regulatory factors that control plant gene expression. It regulates key enzyme genes and transcription factors involved in the synthesis of G. biloba flavonoids[74,75] (Fig. 2). Through high-throughput sequencing, 174 new miRNAs were identified in the leaves of female and male G. biloba leaves. The target of miR1108, gbl-miR174, gbl-miR102, and miR1854 was predicted to be GbFLS1[76]. MiR159a and miR159c in G. biloba are involved in chloroplast development, hormone metabolism, and flavonoid metabolism[77]. Some studies used degradation data to verify the targeting relationship between miRNAs and genes related to flavonoid synthesis. It was found that miR159b targeted Gb4CL6, miRN354 targeted GbANR4, miR167f targeted GbANR7, miRN252 and six members of miRN49 family targeted GbLAR[39]. Non-coding RNAs longer than 200 nucleotides are called long-stranded non-coding RNAs (lncRNAs)[78]. They are involved in the regulation of G. biloba flavonoid synthesis by regulating the key enzyme genes GbPAL, GbCHS, GbCHI, GbF3H, and GbFLS[79]. A total of 14 lncRNAs targeting 16 genes were predicted to be involved in secondary metabolic pathways. In particularly, MSTRG.17450.1 targeted Gb10032, MSTRG.40776.1 targeted Gb25343, MSTRG.5250.2 and MSTRG.18854.1 targeted GbPAL, MSTRG.65322.1, MSTRG.36921.1 and MSTRG.38052.1 targeted GbFLS[80]. LncRNAs can also form a regulatory network with miRNAs and the pyruvate carboxylase gene to regulate the synthesis of G. biloba flavonoids[75]. Sequencing technology is continuously improving, which also improves the analysis of non-coding RNAs that target key genes involved in flavonoid synthesis and metabolism in G. biloba. These improvements will advance our understanding of the regulatory network involved in flavonoid metabolism.
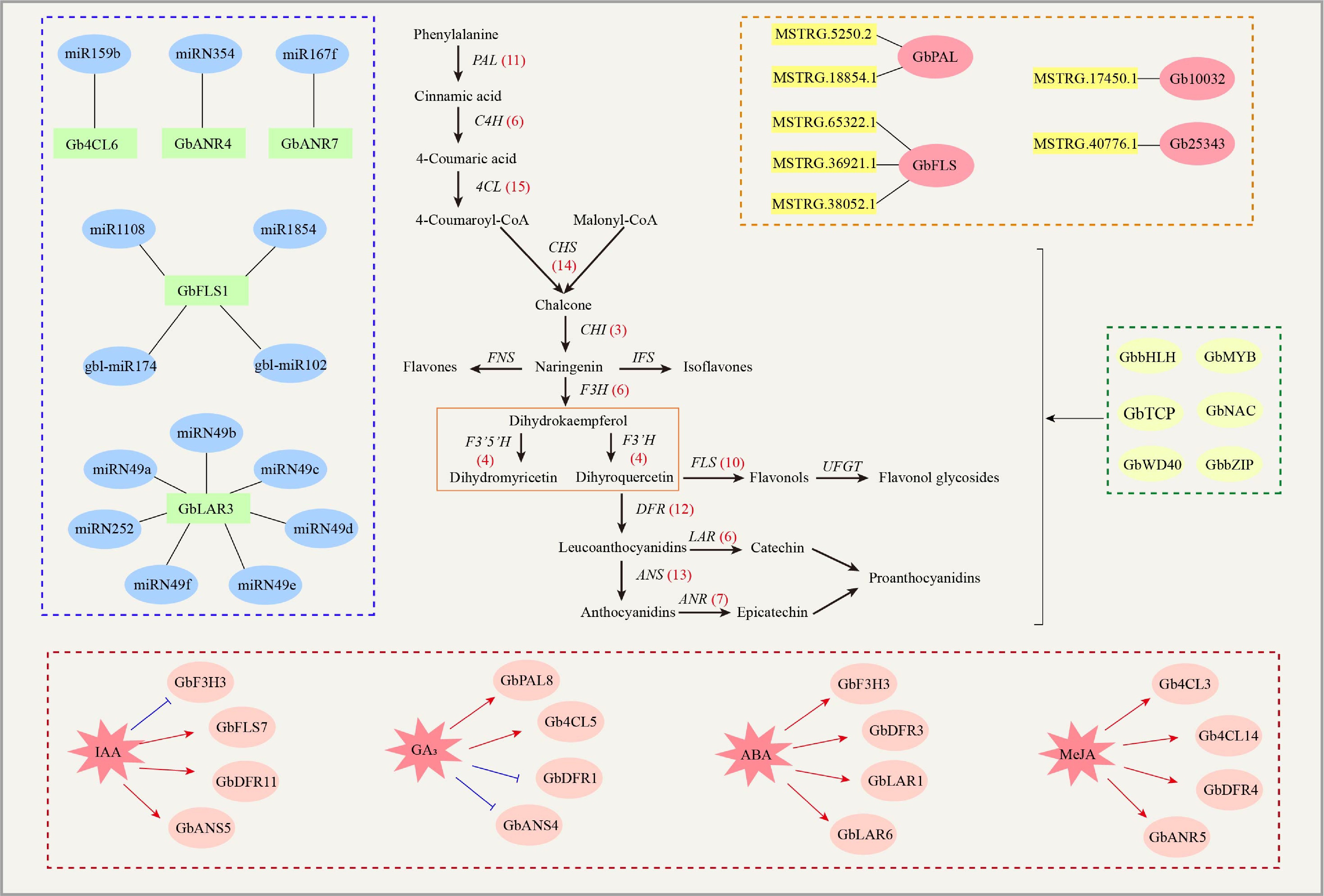
Figure 2.
The regulation network of flavonoids in G. biloba. The synthesis of flavonoids in G. biloba is regulated by several transcription factors including MYB, bZIP, bHLH, WD40, NAC and TCP. In addition, hormones, miRNAs and lncRNAs can also regulate the synthesis of flavonoids in G. biloba by regulating key enzyme genes. The numbers in brackets indicate the number of genes in G. biloba. ANR: Anthocyanidin reductase; ANS: Anthocyanin synthetase; C4H: Cinnamate-4-hydroxylase; CHS: Chalcone synthase; CHI: Chalcone isomerase; 4CL: 4-coumarate: coenzyme A ligase; DFR: Dihydroflavonol 4-reductase; FNS: Flavone synthase; F3H: Flavanone 3-hydroxylase; F3’H: Flavanone 3'-hydroxylase; F3’5’H: Flavanone 3'5'-hydroxylase; FLS: Flavonol synthase; IFS: Isoflavone synthase; LAR: Leucoanthocyanidin reductase; PAL: Phenylalanine ammonia-lyase; UFGT: UDP glucose: flavonoid 3-O-glucosyltransferase.
-
In this review, we have summarized the current understanding of flavonoid function and biosynthesis in the important medical plant G. biloba. Flavonoids are important secondary metabolites with high medicinal value. G. biloba is rich in flavonoids, and flavonoids are distributed in almost all the organs, including the leaf, seed, bud, stem, and embryo. Most of the G. biloba flavonoids exist in the form of glycosides and mainly consist of monoflavones (e.g., kaempferol, quercetin, and isorhamnetin) and biflavones (e.g., ginkgetin, isoginkgetin, sciadopitysin, and bilobetin). The last few decades have witnessed the progress of various roles of G. biloba flavonoids in resisting stress. For example, they can absorb UV-B to protect against ultraviolet radiation. Under moderate drought stress, oxidizing substances (ROS) accumulated to promote the accumulation of flavonoids, thus enhancing the ability to resist drought. On the other hand, G. biloba flavonoids can fight influenza viruses by inhibiting the activity of sialidase. They induce S-phase arrest in the intracellular environment to defend against fungi. In particular, they can inhibit changes in general metabolic parameters such as body weight, fat mass, insulin level, and glucose tolerance activity to prevent cardiovascular dysfunction, which is of great benefit to human health.
It is noteworthy that several factors affect flavonoid production in G. biloba leaves. Flavonoid content in the leaves of young trees is much higher than that of adult trees; thus, only the leaves of young trees are qualified as raw materials for drugs in production, indicating that the age of trees plays an important role in the accumulation of flavonoids in G. biloba leaves. Therefore, it is of great significance to study the regulation mechanisms of environmental factors and age (or rejuvenation) on flavonoid synthesis in G. biloba in the future.
The successful assembly of the G. biloba genome provides data resources for further study on the function and regulation mechanisms of key genes in flavonoid synthesis. Combined with multi-omics analysis, a series of functional genes regulating flavonoid synthesis were identified and found to be extensively expanded in G. biloba. In addition, several transcription factors were predicted to be involved in flavonoid synthesis, such as MYB, bHLH, HY5, and WD40, and degradome analysis showed that multiple miRNAs may target these genes. Although these genes have been predicted to be involved in transcription and post-translational regulation of flavonoid synthesis, their function and regulatory mechanisms are still unclear due to the lack of G. biloba transformation system. Therefore, it is urgent and necessary to establish the genetic transformation system in G. biloba, such as root hair transformation, VIGS transformation and gene editing systems (CRISPR-Cas9) to verify their function, as well as apply the molecular biology technologies, such as Y1H, EMSA and dual luciferase assay to explore the regulatory mechanism in flavonoid synthesis. The development of these technologies in G. biloba will provide a platform for the identification of key genes and regulatory modules or networks that regulate the synthesis of flavonoid. Unveiling fundamental genetic knowledge toward high flavonoid biosynthesis will be essential for improving efficient cultivation and precise breeding of G. biloba in the future.
This work was supported by the National Natural Science Foundation of China (Grant No. 31971686 and 32171838), Forestry Sci-tech Innovation and Promotion Project of Jiangsu Province (LYKJ[2021]35), Jiangsu Agriculture Science and Technology Innovation Fund (Grant No. CX(21)3047).
-
The authors declare that they have no conflict of interest.
- Copyright: © 2023 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Mao D, Zhong L, Zhao X, Wang L. 2023. Function, biosynthesis, and regulation mechanisms of flavonoids in Ginkgo biloba. Fruit Research 3:18 doi: 10.48130/FruRes-2023-0018
Function, biosynthesis, and regulation mechanisms of flavonoids in Ginkgo biloba
- Received: 06 February 2023
- Accepted: 09 June 2023
- Published online: 03 August 2023
Abstract: Ginkgo biloba is an important economic tree species. Due to the abundance of secondary metabolites in the tree, Ginkgo biloba extract (GbE) is used as a medicine, food supplement, and nutraceutical. Flavonoids are the most active components in GbE. There is increasing evidence that external and internal factors affect flavonoid synthesis. The publication of the G. biloba genome significantly improved functional analyses of key genes, transcription factors, and non-coding RNAs involved in flavonoid synthesis. Here, we review progress on understanding the mechanisms of external and internal factors that affect the synthesis and accumulation of flavonoids in G. biloba. We highlight recent achievements that have greatly advanced our understanding of the functions of key genes for flavonoid synthesis. In addition, we discuss novel insights into the metabolic regulation network of flavonoids in G. biloba.
-
Key words:
- Ginkgo biloba /
- Flavonoid accumulation /
- Biosynthesis /
- Gene regulation












