-
MADS-box transcription factors (TFs), which incorporate an evolutionarily conserved domain (MADs), can be found across a diverse array of eukaryotes[1]. Land plants contain a higher proportion of MIKC*- and MIKCC-type MADS-box TFs than any other group of organisms[2,3]. Theoretically, the accumulation of MADS-box genes in angiosperms is likely to have facilitated the morphological diversity exhibited by this group of plants. The 'ABC' model of floral organogenesis was proposed following genetic studies in Antirrhinum majus and Arabidopsis thaliana[4]. The model was later revised to the 'ABC(D)E' model, after the addition of D and E class and MIKCC genes[5,6]. In general, it is believed that MADS-box proteins act synergistically during the process of primordial floral organogenesis: C + D + E class genes specify ovules; C + E class genes specify carpels; B + C + E class genes specify stamens; A + B + E class genes specify petals; and A + E class genes specify sepals[7,8]. Several genes related to floral organogenesis have been characterized in Arabidopsis, including the E class genes SEPALLATA1-4 (SEP1-4), the C class gene AGAMOUS (AG), the B class genes APETALA3 (AP3) and PISTILLATA (PI), and the A class genes APETALA1 (AP1) and APETALA1 (AP2). Each of these genes is considered a MIKCC MADs-box genes, except for AP2.
Two paralogous MADS-box gene lineages, PISTILLATA (PI) and APETALA3 (AP3), are likely to have arisen before the rise of the angiosperms during a duplication event. In both Arabidopsis and snapdragon, in the third whorl, these genes determine stamen identity and in the second whorl they determine petal identity[9,10]. Across core eudicots and monocots, the expression of B class genes is relatively conserved in stamens, petals, or lodicules. However, divergent expression of B class genes outside of the core eudicots is associated with poorly differentiated perianth. Novel floral morphologies are often associated with B class gene diversification in both function and expression. In Petunia for example, stamen filament and petal tube fusion is regulated by GLO1[11]. Staminodia, novel floral organs in Aquilegia, appear to be specified by B class genes[12]. In Magnoliids, Ranunculales, and basal angiosperms and monocots, homeotic petaloid organogenesis is affected by B class MADS box gene ectopic expression. The expression patterns of B homologous genes from basal eudicots Akebia trifoliata, Platanus acerifolia and Trochodendron aralioides have been investigated deeply in the developing tissues, indicating that divergent function probably involved floral organ identity[13−15].
As with the duplication event resulting in the establishment of the PI and AP3 genes, a whole genome duplication (WGD) event occurring before the rise of the angiosperms likely contributed to the AGL11 and AG gene lineages, previously defined as C and D lineages[16,17]. In Arabidopsis, the C class gene AGAMOUS (AG) is responsible for carpel and stamen organogenesis, determination of floral meristem fate, and prevention of A class gene misexpression in floral tissues[18]. In addition to C class gene function, the AGL11-like gene STK exhibits a particular function in ovular funiculus development[19,20]. In the basal eudicot Papaver somniferum, stamen and carpel features are determined by an AG orthologue, with AG-1 involved in stamen and carpels AG-2 involved only in carpels[21].
Because Epimedium plants have evolved unique floral morphologies, including petaloid sepals and spur or sac-like petals with nectariferous tissue on their inner faces[22], these plants are ideal investigation models. Previous studies primarily characterized the function of the Crabs claw gene in the development of nectaries and the AP1/SEP/AGL6 MADS-box superclade in flower development and flavonoid synthesis[19,23−25]. Here, we have functionally characterized the AP3, PI, AG, and AG11 homologs of the basal eudicot Epimedium sagittatum (Berberidaceae, Ranunculales). The results of this study will provide a clearer understanding of how C and B class genes are regulated in E. sagittatum.
-
Five MADS-box genes were discovered in the floral tissues of E. sagittatum. Through a BLAST search against the NCBI database, these genes were originally assigned to the C and B class MADS-box genes. MEGA5 was utilized to perform a phylogenetic characterization of the AG11/AG and PI/AP3 genes from E. sagittatum and other plants using the maximum likelihood method with MEGA5 software[26]. EsAP3-1, EsAP3-2, and EsAP3-3 were found to belong to the previously characterized and evolutionarily conserved AP3-III, AP3-II, and AP3-I paralogous lineages (Fig. 1). In contrast, no duplication events were found for the PI-like gene (Fig. 2). Based on their topology, EsAG and EsAG11 were assigned to the AG and AG11 lineages (Fig. 3).
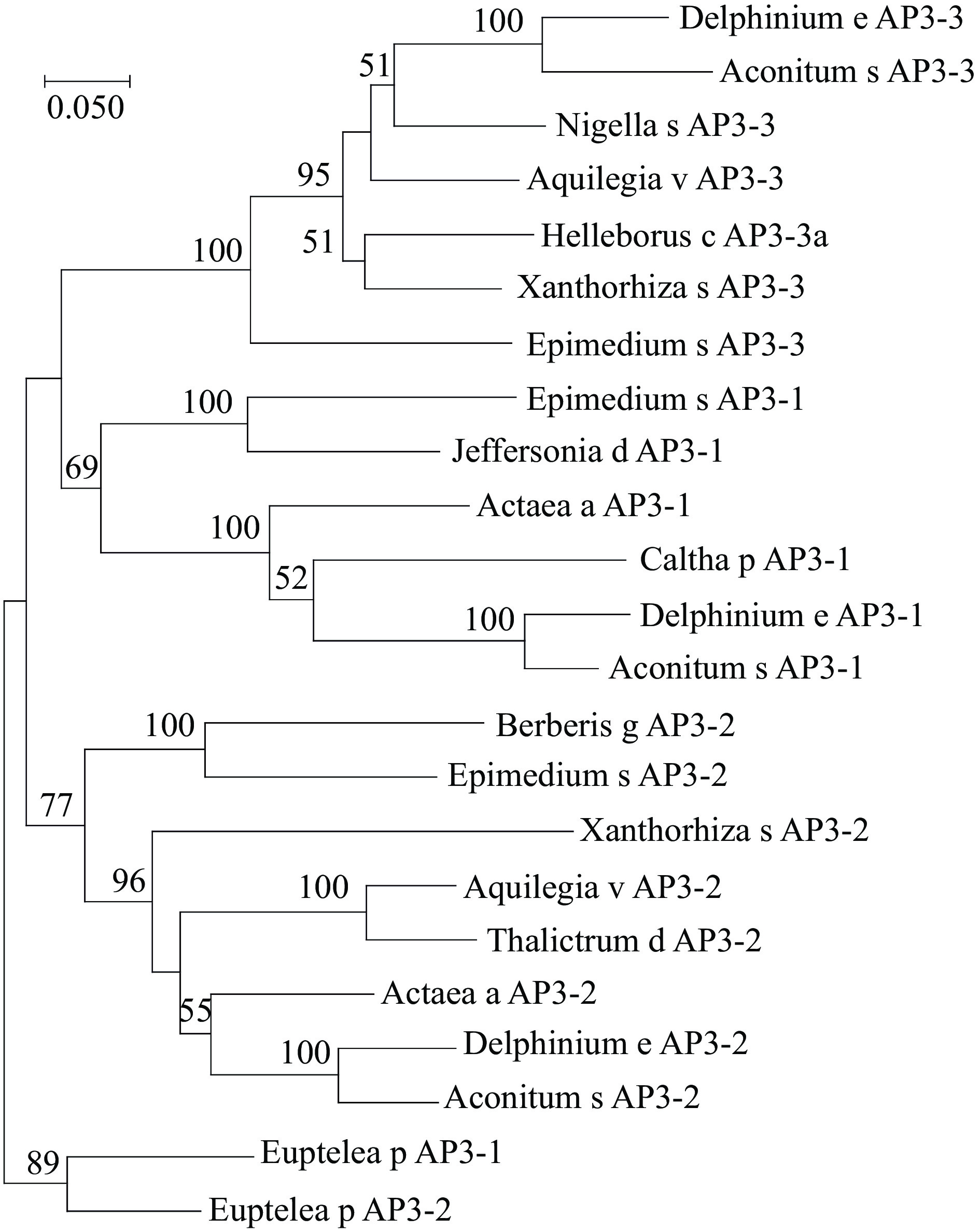
Figure 1.
Phylogeny of AP3 lineage genes. The maximum likelihood tree was produced using MEGA5 from multiple nucleotide alignments and reveals that the phylogenetic position of EsAP3-1, EsAP3-2 and EsAP3-3. Bootstrap values over 50 was shown with 1000 replications. Caltha p AP3-1, EU481813.1; Actaea a AP3-1, HQ647375.1; Delphinium e AP3-1, EU481804.1; Aconitum s AP3-1, EU481818.1; Jeffersonia d AP3-1, EU481791.1; Xanthorhiza s AP3-2, EU481797.1; Delphinium e AP3-3, EU481802.1; Aconitum s AP3-3, EU481816; Nigella s AP3-3, HQ694794.1; Helleborus c AP3-3a AY162876.1; Aquilegia v AP3-3, EF489476.1; Xanthorhiza s AP3-3, EU481796.1; Berberis g AP3-2 AY162859.1; Euptelea p AP3-1, GU357449.1; Euptelea p AP3-2, GU357450.1; Aquilegia v AP3-2, EF489477.1; Thalictrum d AP3-2, AY867876.1; Actaea a AP3-2 HQ647376.1; Delphinium e AP3-2, EU481803.1; Aconitum s AP3-2, EU481817.1.
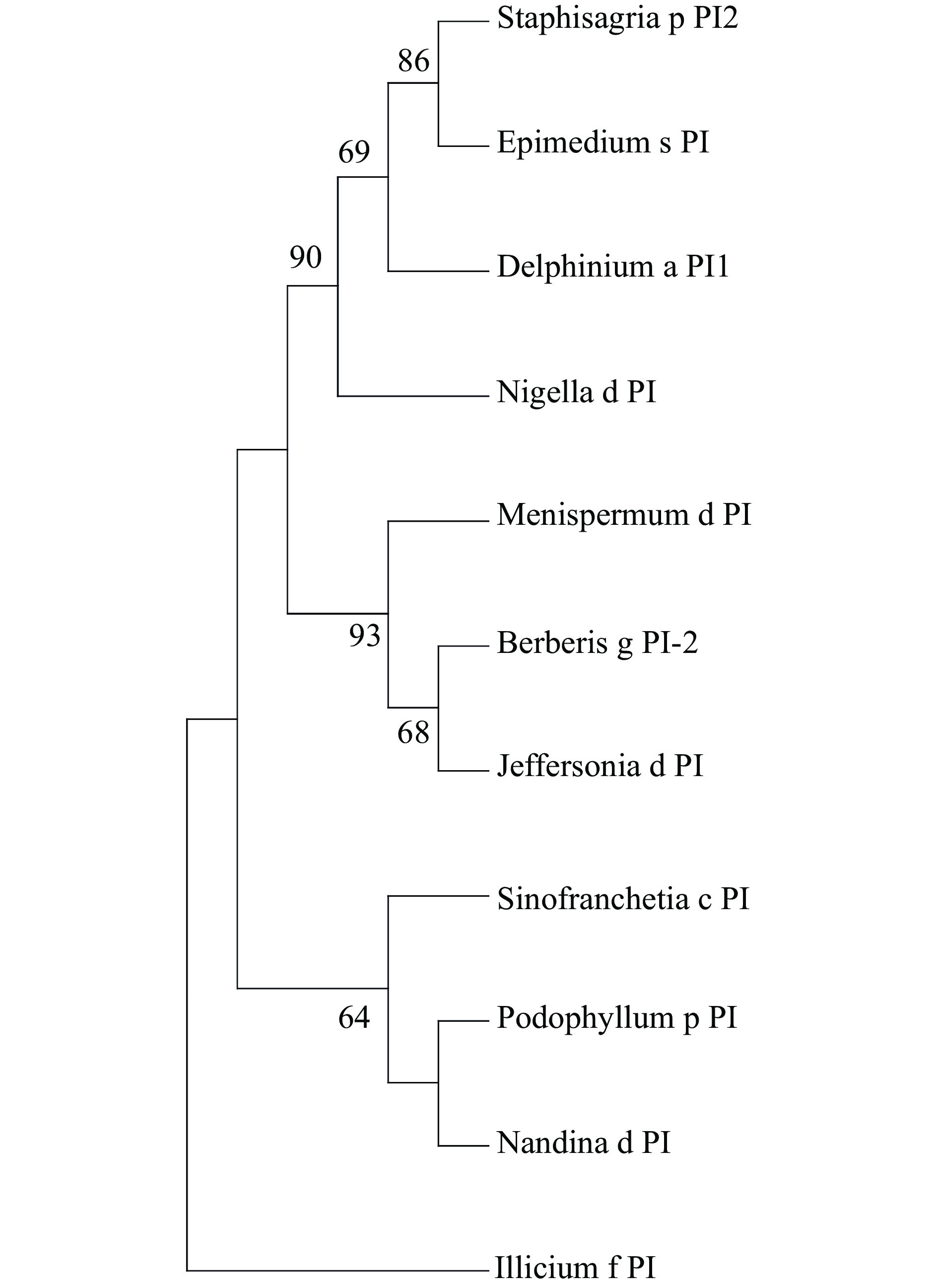
Figure 2.
Phylogeny of PI lineage genes. The ML phylogenetic tree from multiple nucleotide alignments of PI homologs from E. saggitatum and other species was conducted by MEGA software (version 5). The PI sequence from Illicium floridanum was used as an outgroup. Bootstrap test was estimated with 1000 replicate analysis, above 50 support was shown. Genbank Accession Numbers: Podophyllum p PI, HQ694792; Sinofranchetia c PI JQ806397; Staphisagria p PI2 OL469304; Berberis g PI-2 AY162861; Jeffersonia d PI EU481792; Nigella d PI KT984414;Delphinium a PI1 OL469301; Nandina d PI HQ694793; Menispermum d PI EU481787; Illicium f PI AY936224.
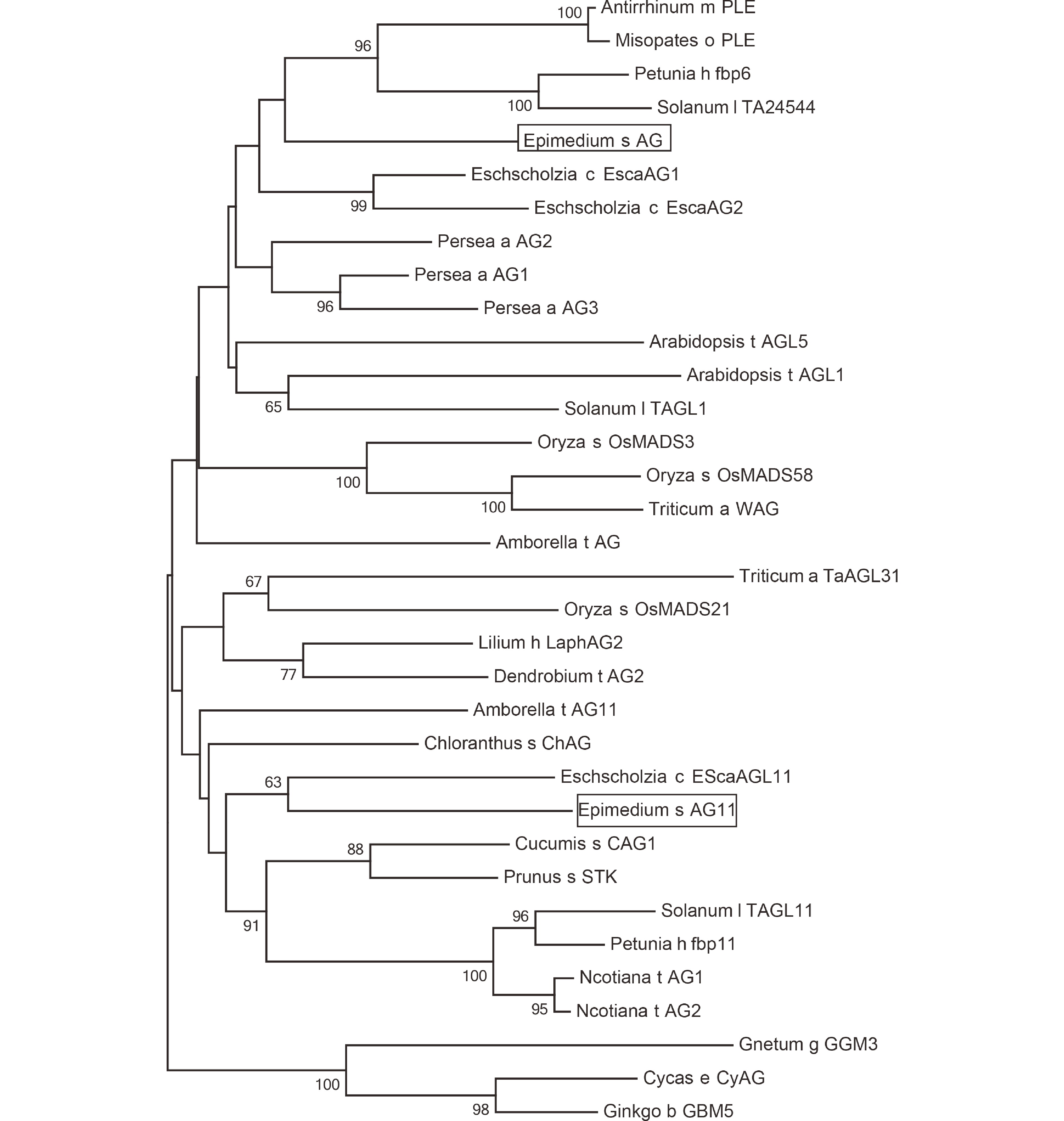
Figure 3.
Phylogeny of AG and AG11 lineage genes from homologs from E. sagittatum and other angiosperm species. The C class genes from Gnetum gnemon, Cycas edentata and Ginkgo biloba sequences were used as an outgroup. The ML phylogenetics tree was constructed by MEGA (version 5). Bootstrap test was estimated with 1000 replicate analysis, above 50 support was shown. Genbank Accession Numbers: Arabidopsis t AGL5, M55553.1; Triticum a TaAGL31, DQ512349.1; Oryza s OsMADS21, FJ750944.1; Arabidopsis t AGL1 NM_118013.2; Eschscholzia c EScaAGL11, DQ088998.1; Lilium h LaphAG2, AB359184.1; Dendrobium t AG2, DQ017703.1; Amborella t AG11, XM_006858527.2; Chloranthus s ChAG, AY464099.1; Solanum l TAGL11, AY098736.2; Nicotiana t AG1, KM205819.1; Nicotiana t AG2, NM_001325570.1; Petunia h fbp11, X81852.1; Cucumis s CAG1, NM_001280577.1; Prunus s STK, GU332504.1; Gnetum g GGM3, AJ132209.1; Oryza s OsMADS3, L37528.1; Oryza s OsMADS58, AB232157.1; Triticum a WAG, AB084577.1; Cycas e CyAG, AF492455.1; Ginkgo b GBM5, GU563899.1; Antirrhinum m PLE, AB516404.1; Misopates o PLE, AM162212.1; Petunia h fbp6, X68675.1; Solanum l TA24544, AY098735.2; Amborella t AG, NM_00 1305835.1; Eschscholzia c EscaAG1, DQ088996.1; Eschscholzia c EscaAG2, DQ088997.1; Persea a AG2, DQ398011.1; Persea a AG1, DQ398021.1; Persea a AG3, DQ398023.1.
Patterns of expression of E. sagittatum floral MADS-box genes
-
Gene expression of both C and B class MADS-box genes in mature floral tissue of E. sagittatum was evaluated using qRT-PCR (Fig. 4). We combined stamens and carpels due to the difficulty of separating them. Consistent with previous studies on Epimedium, EsAP3-3 expression was restricted to floral nectar spurs. Both EsAP3-1 and EsAP3-2 were expressed in leaves, roots, and flowers. EsPI exhibited high expression in petals, but displayed minimal expression in stamens/carpels and sepals. EsAG was expressed in stamens/carpels, petals, and sepals, while EsAG11 exhibited stamen/carpel-specific expression.
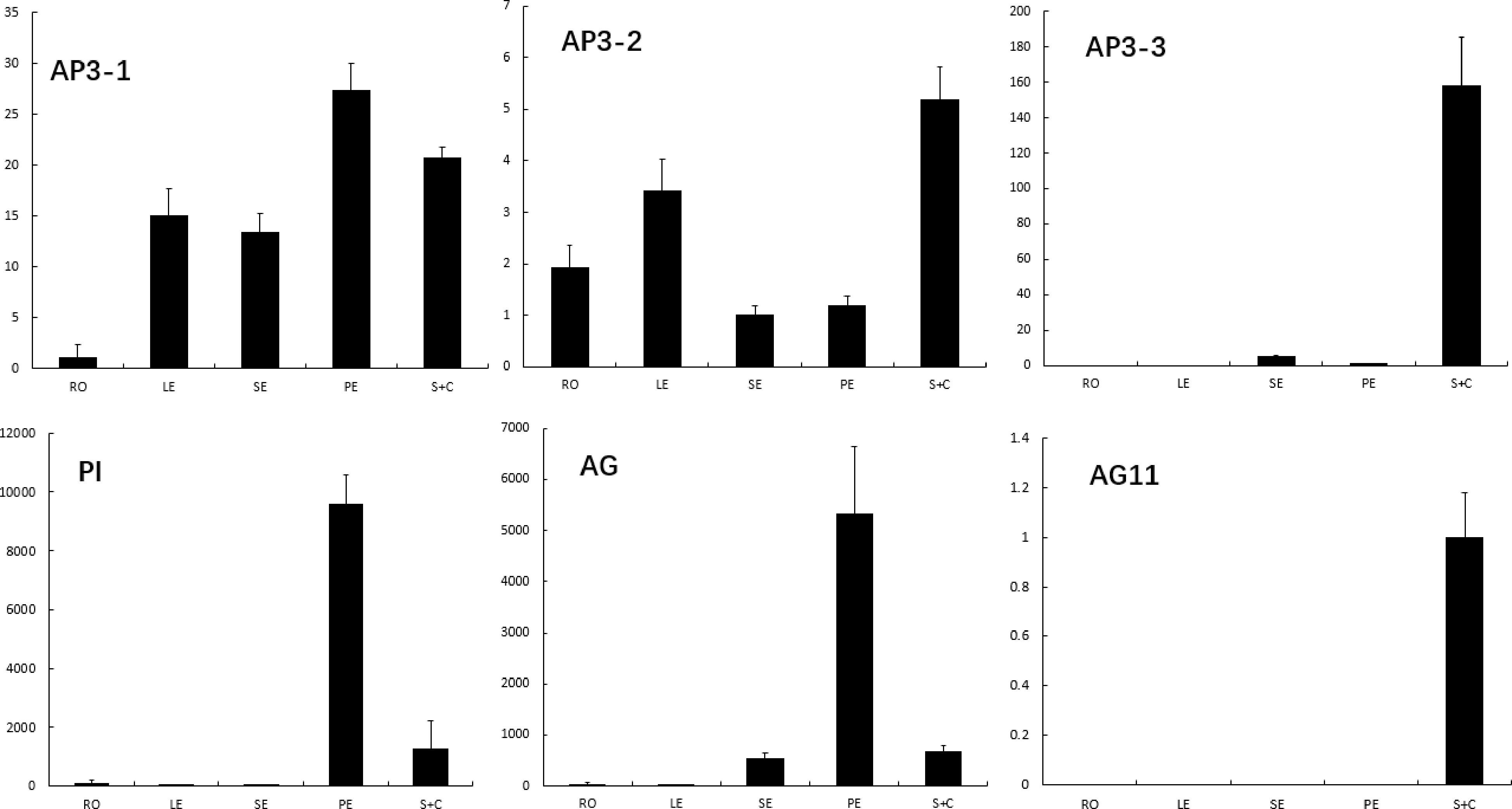
Figure 4.
The expression of E. sagittatum B and C class genes in root, leaf, seqal, petal and combination of stamen and carpel. Error bars represent SE for three technical replicates. The expression of each gene was normalized against the Epimedium actin gene.
Interaction analysis of the E. sagittatum C and B class proteins
-
Interactions between C and B class proteins were probed using a yeast-two-hybrid assay. Self-activation was not observed for any of the tested proteins after fusion to the GAL4 domain. Additionally, B class proteins exhibited no interactions with C class proteins, and vice versa. Compared with other B class proteins, EsAP3-1 has broadly interactive partners. Furthermore, EsPI may be able to form a heterodimer with EsAP3-1 and EsAP3-2. Additionally, homodimer and semihomodimer interactions were observed between EsAP3-1, EsAP3-2, and EsAP3-3 (Table 1).
Table 1. Data from yeast two-hybrid assay. EsAP3-1, EsAP3-2, EsAP3-3, and EsPI were added to both an activation and GAL4 binding domains. Interactions were analyzed using SD/-Ade/-His/-Trp/-Leu plates containing 10 mM of 3-amino-1,2,4-triazole. Three replicates were analyzed for each experiment.
BD-EsAP3-1 BD-EsAP3-2 BD-EsAP3-3 BD-EsPI BD-EsAG BD-EsAG11 AD-EsAP3-1 + + + + − − AD-EsAP3-2 + − + − − − AD-EsAP3-3 − − − − − − AD-EsPI + − +/− − − − AD-EsAG − − − − − − AD-EsAG11 − − − − − − Multimeric complex analysis C and B class proteins
-
The ability of E. sagittatum B and C class MADs-box proteins to organize into synergistic complexes was explored in yeast. As previously reported, neither EsAG nor EsAG11 were observed to organize into a heterodimer with the B class proteins. However, AP3-2 (AP3-2-IKC) appears to be able to bring either EsAG or EsAG11 and EsPI together to form a complex (Table 2). In addition, EsAG11 and EsPI interacted strongly when co-expressed together with either AP3-3 (AP3-3-IKC) or PI (PI-IKC) sans MADS-box.
Table 2. Data from yeast three-hybrid assay examining the formation of higher order protein complexes.
Ternary complex -LTAH + 10 mM 3AT,
28C and LacZiAD-EsPI + pTFT-AP3-1ΔM + BD − AD-EsPI + pTFT-AP3-2ΔM + BD − AD-EsPI + pTFT-AP3-3ΔM + BD − AD-EsPI + pTFT-PIΔM + BD − AD-EsPI + pTFT-AP3-1ΔM + BD-EsAG − AD-EsPI + pTFT-AP3-1ΔM + BD-EsAG11 − AD-EsPI + pTFT-AP3-2ΔM + BD-EsAG + AD-EsPI + pTFT-AP3-2ΔM + BD-EsAG11 + AD-EsPI + pTFT-AP3-3ΔM + BD-EsAG − AD-EsPI + pTFT-AP3-3ΔM + BD-EsAG11 + AD-EsPI + pTFT-PIΔM + BD-EsAG − AD-EsPI + pTFT-PIΔM + BD-EsAG11 + AD + pTFT-AP3-1ΔM + BD-EsAG − AD + pTFT-AP3-1ΔM + BD-EsAG11 − AD + pTFT-AP3-2ΔM + BD-EsAG − AD + pTFT-AP3-2ΔM + BD-EsAG11 − AD + pTFT-AP3-3ΔM + BD-EsAG − AD + pTFT-AP3-3ΔM + BD-EsAG11 − AD + pTFT-PIΔM + BD-EsAG11 − AD + pTFT-PIΔM + BD-EsAG11 − Functional analysis of EsAG through Arabidopsis transformation
-
Agrobacterium-mediated transformation of wild-type Arabidopsis was used for functional characterization of EsAG. Specifically, the EsAG gene was cloned into ectopic expression vectors containing the CaMV 35S promoter, and four transgenic 35S::EsAG Arabidopsis lines were obtained. The transformed lines were subsequently confirmed by qRT-PCR (Fig. 5a). A few of the transgenic plants exhibited distinct phenotypes (Fig. 5b), and sepals were homeotically transformed into stigmatic papillae-containing carpelloid structures in other lines (Fig. 5c).
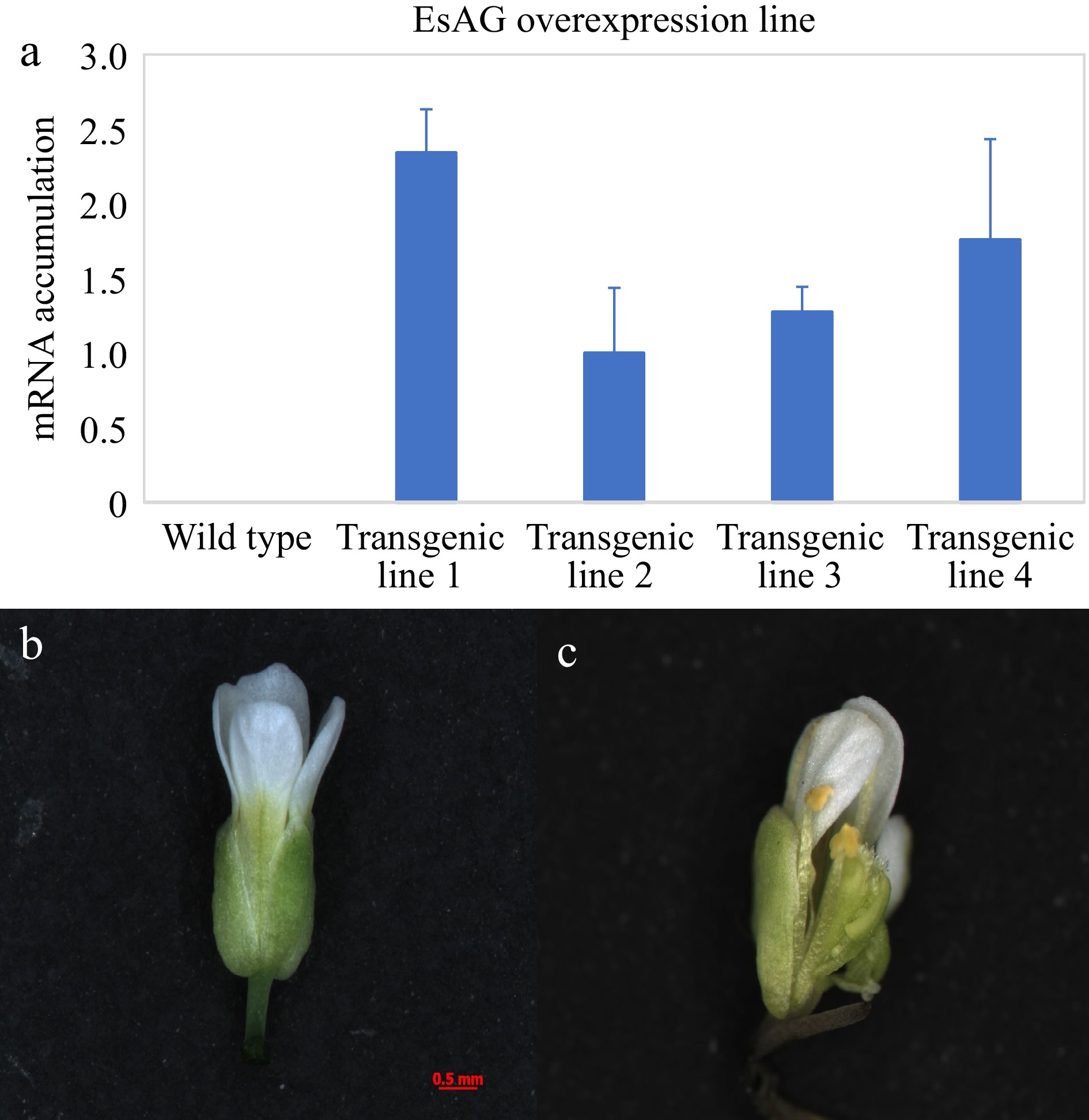
Figure 5.
Functional analysis of EsAG in wild-type Arabidopsis. (a) Analysis of EsAG overexpression lines in 35S::EsAG transgenic Arabidopsis plants. A fragment of Tubulin gene was amplified as an internal control. Error bars represent SE for three technical replicate reactions. (b) The flower of the wild-type Arabidopsis. (c) Phenotype of 35S::EsAG transgenic Arabidopsis in the wild-type background.
-
To study their roles in the regulation of floral organogenesis, we identified and characterized four B class MADS-box genes (EsAP3-3, EsAP3-1, EsAP3-2, and EsPI) through motif and phylogenetic analyses. We found that the three AP3-like genes belonged to three phylogenetic lineages, consistent with previous reports suggesting that, prior to the divergence of Ranunculaceae and Berberidaceae, two WGD events occurred[27]. Furthermore, gene duplication events resulting in alterations in gene expression or coding regions occurred at different scales after plant polyploidy.
EsAP3-1 and EsAP3-2 exhibited broad expression across floral tissues, although EsAP3-3 exhibited petal-specific expression, which is consistent with data suggesting that the expression patterns AP3-3 orthologs among Ranunculales is conserved[27−30]. The absence of AP3-3 paralog expression in Aquilegia vulgia and Nigella damascene is associated with the loss of petal initiation[29]. Functional validation will be required to fully characterize the role of AP3-3 paralogs in petal formation in Epimedium. Furthermore, differential heterodimerization patterns among AP3-3 paralogs is implicated in the diverse roles these genes play in floral organ formation. Our study of the AP3-PI and C protein complex suggests that all three E. sagittatum AP3-like genes are likely to have been subfunctionalized after WGD, similarly to EsAP3-I and EsAP3-2.
We found two novel conserved AG motifs (motifs II and I) in the EsAG and EsAG11 C-terminal regions, which is consistent with previous studies[16,31]. These conserved motifs are likely related to the core eudicot duplication event which produced the AG and AG11 lineages. As with other D and C class genes, both EsAG and EsAG11 exhibit floral tissue-specific expression patterns, where petal and stamen formation are controlled by these orthologs. In Arabidopsis, AG orthologs regulate carpel and stamen organogenesis dictate ovule identity though synergistic interactions with E and D class genes, limit the determinacy of floral meristematic tissue, and act as repressors of A class genes[4] . The ability of both EsAG and EsAG11 to differentially interact with the B protein dimer complex in E. sagittatum indicates the possibility that they work to regulate floral organogenesis. Transgenic overexpression of AG in Arabidopsis caused sepals to homeotically transform into carpelloid structures, and petals to transform into stamen-like structures[32]. Here, we also observed phenotypic alterations in the EsAG transgenic Arabidopsis line, suggesting that EsAG regulates carpel and stamen organogenesis across plant lineages.
-
Field-grown E. sagittatum transplants were cultivated at the Wuhan Botanical Garden at the Chinese Academy of Sciences (Wuhan, China). Columbia ecotype Arabidopsis thaliana plants were grown using 8 h of dark and 16 h of light.
Cloning of C and B class genes from E. sagittatum
-
Leaves, roots, and inflorescences of E. sagittatum were used to extract total RNA with the Trizol reagent (Invitrogen, CA, USA), and subsequently digested using RQ1 RNase-free DNase (Promega, WI, USA). First-strand cDNA was transcribed with and the poly-T primer 3'-CDS and Superscript III reverse transcriptase (Invitrogen, CA, USA). 5'- and 3'-RACE PCR were used to isolate the EsAP3-I, II, III, EsPI, EsAG, and EsAG11 genes. 3'-RACE was performed using the p5B and SMARTII primers for B class genes, and the B2 primer for EsAG and EsAG11. First strand synthesis was carried out with the 5'-CDS and SMART II primers. Two 5' RACE reactions were carried out using the 5' specific primer 1 and UPM for the first strand, and 5' specific primer 2 and NUP for the second strand. Both the 3’ and 5' UTRs were utilized to generate full-length sequences. The primers used for this experiment can be found in Supplemental Table S1.
qRT-PCR
-
The expression of EsAP3-I, EsAP3-II, EsAP3-III, EsPI, EsAG, and EsAG11 were investigated in leaves and floral organs, including carpels, stamens, petals, and sepals, at both preanthesis and anthesis. Prior to qRT-PCR, a PrimeScript RT kit (Takara, Japan) was utilized to perform first strand synthesis and DNA decontamination. All assays were performed using a 20 μl reaction volume, including 10 ng of cDNA, and 10 μM SYBR Premix Ex Taq II and primers (Takara, Japan). qRT-PCR was performed with an ABI7500 Real-Time PCR system (ABI, USA), using the following protocol: denaturation stage (30 s at 95 °C, then 40 cycles of 3 s at 94 °C and 30 s at 60 °C) and dissociation stage (15 s at 95 °C, 1 min at 60 °C, 30 s at 60 °C). Gene expression was quantified using the two-delta CT relative calculation method. EsActin was used as the internal reference. Each experiment consisted of three technical replicates for each of three biological replicates. Data was handled using software provided in ABI7500.
Phylogenetic analysis
-
Phylogenetic analyses of specific C and B class genes were performed on full-length nucleotide alignments. The maximum likelihood (ML) method was employed in MEGA5 with the GTR model and four categories of rate substitution. ML analyses were carried out using 500 bootstrap replications and the default settings. Data were used to estimate the Gamma distribution. The tree topology was obtained through the nearest-neighbor interchange strategy.
Yeast three- and two-hybrid assays
-
EsAG and EsAG11 full-length genes together with the IKC parts from full-length EsAP3-I, EsAP3-II, EsAP3-III, and EsPI were amplified from pMD19-T using primers that introduced 3’ and 5’ restriction sites. The resultant PCR products were subsequently digested using the corresponding restriction enzymes and introduced into the pGADT7 and pGBKT7 vectors (Clontech, CA, USA). For the yeast three-hybrid assay, the third protein, containing both the ADE selectable marker gene and the full-length ADH1 promoter, was expressed from the pTFT1 vector. M-domain lacking EsAP3-I, EsAP3-II, EsAP3-III, and EsPI were cloned, in frame with the SV40 coding region, into the pTFT1 vector. Negative controls consisted of co-transformed AD or BD empty and insertion vectors. For the yeast two-hybrid assay, the constructed empty pGADT7 and pGBKT7 were introduced into AH109 yeast (Saccharomyces cerevisiae) according to the LiAc transformation procedure to detect autoactivation. Yeast three- and two-hybrid assays were tested by using SD medium lacking tryptophan, histidine, adenine, and leucine, and containing 10 mM 3-Amino-1, 2, 4-triazole.
Transgenic analysis
-
In order to construct the p35Spro EsAG vector, KpnI and SalI were utilized to cut the full-length versions of each gene from the PMD19-T vector (Takara, Japan). The genes were then inserted into the pMV binary vector (derived from pBI121) after previous digestion with KpnI and XhoI. Agrobacterium tumefaciens GV3101 was used to introduce the construct into Arabidopsis through floral dipping[33]. Murashige and Skoog medium at half strength containing 50 ug/mL Kanacymin was used to select transgenic seedlings. Transgenic leaf-level gene expression was determined using qRT-PCR, with Tublin as the internal reference gene.
We thank Drs Elena Kramer and Hongzhi Kong for communications on MADS-box gene family, Dr Richard Immink and for yeast experimental assistance. This work was supported by National Key R&D Program of China (2022YFC3501703); the Scientific and Technological Innovation Project of China Academy of Chinese Medical Sciences (C12021A03710);
-
Wei Sun is the Editorial Board member of journal Medicinal Plant Biology. He was blinded from reviewing or making decisions on the manuscript. The article was subject to the journal's standard procedures, with peer-review handled independently of this Editorial Board member and his research group.
-
# Authors contributed equally: Wei Sun, Huihua Wan
- Supplemental Table S1 All primer sequences used for the experiments.
- Copyright: © 2023 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Sun W, Wan H, Huang W, Yousaf Z, Huang H, et al. 2023. Characterization of B- and C-class MADS-box genes in medicinal plant Epimedium sagittatum. Medicinal Plant Biology 2:1 doi: 10.48130/MPB-2023-0001
Characterization of B- and C-class MADS-box genes in medicinal plant Epimedium sagittatum
- Received: 18 October 2022
- Accepted: 30 January 2023
- Published online: 27 February 2023
Abstract: The basal eudicot Epimedium (Barrenwort) exhibits innovative floral morphology in the form of petal spurs filled with nectar and petaloid sepals. The B-class MADS-box genes APETALAS3 (AP3) and PISTILLA (PI) determine sepal and petal identity while the C-class gene AGAMOUS (AG) determines carpel and stamen identity in Arabidopsis. Complex histories of gene duplication resulted in subsequent subfunctionalization or neofunctionalization of paralogs. Here, a total of four B- and two C-class genes were successfully isolated from E. sagittatum. Phylogenetic analysis showed that EsAP3-1, EsAP3-2, and EsAP3-3 are part of the AP3 clade; EsPI is part of the PI group; and EsAG and EsAG11 clustered into the AG and AG11 groups, respectively. Quantitative real-time PCR was utilized to detect the expression patterns of these genes, and all B-class genes except for EsAP3-3 were found to be universally expressed. The transcribed EsAG and EsAG11 genes were confined to reproductive organs. In addition, yeast three and two hybrid assays were used to explore the status of protein complexes. EsAP3-1 was found to have broadly interactive partners, and EsPI can form a heterodimer with EsAP3-1 and EsAP3-2. Transgenic EsAG overexpression in wild-type Arabidopsis confirmed the conserved function determining carpel development.
-
Key words:
- Epimedium /
- MADs-box /
- AP3/PI /
- Medicinal plant













