-
Sugarcane (Saccharum officinarum) is one of the most important economic crops in tropical and subtropical countries as it is the main raw material for sugar production. Brazil and India are the top two sugarcane producers among the more than 100 countries that grow sugarcane[1,2]. Sugarcane provides approximately 80% of the world’s sugar and more than 90% of the sugar in China[3−5]. Moreover, it has now become a raw material of bioethanol[6]. Sugarcane production is threatened by biotic and abiotic stresses[7−9]. Meanwhile, numerous other characteristics, such as cane yield and other agronomic traits, of commercial and cultivated sugarcane varieties need to be improved by sugarcane scientists to fully meet the needs of sugarcane farmers[10]. The cultivated varieties of sugarcane are the progenies of crosses among Saccharum officinarum, Saccharum spontaneum, Saccharum barberi, and Saccharum sinense[11−14]. They are heterozygous aneuploid crops that contain numerous chromosomes (40–200), and the structure of their genomes is very complex[15]. However, sugarcane is photoperiod-sensitive, and numerous parent germplasms are difficult to cross; all of these factors complicate character improvement through traditional breeding strategies[16]. Genetic transformation is a convenient and effective strategy for the character improvement of cultivated varieties[17]. Brazil is the first and only country utilizing transgenic sugarcane varieties in field production[18]. In 2022–2023, Brazil planted almost 70,000 hectares of transgenic sugarcane, accounting for 89% of annual production. No other country apart from Brazil uses transgenic sugarcane varieties in field production primarily because transgenic sugarcane lines are difficult to obtain in the laboratory[19]. The primary technical difficulty is not easy to obtain high-quality embryogenic calluses from different varieties. The embryogenic calluses of sugarcane are the only explants that can be used for genetic transformation through Agrobacterium-mediated and particle bombardment methods. However, embryogenic calluses and the most suitable status for genetic transformation must be precisely identified. Furthermore, plant physiology and anatomy must be organically combined. Different varieties also need different recipes of callus induction media and different subculture protocols during callus induction. In brief, considerable practice and comparison, time, and resources are needed. In our previous research, three kinds of sugarcane genetic transformation selection systems, namely, the PMI/mannose selection system[20], bar/Basta selection system[21], and CP4-EPSPS/Roundup (unpublished) selection system, were successfully established by enhancing the quality of sugarcane embryogenic callus induction by using Agrobacterium-mediated genetic transformation and a precise transformation protocol. The bar/Basta selection system was the most efficient genetic transformation system for sugarcane. Herein, the key points/steps, advantage, and contribution in China of the efficient bar/Basta transgenic selection system are presented and discussed.
-
The embryogenic callus is an ideal material for transgenesis because of its stable proliferation; high regeneration rate; sensitivity to screening antibiotics and herbicides, such as kanamycin, hygromycin, and glufosinate ammonium; and tolerance to Agrobacterium tumefaciens[22−24]. The low quality of embryogenic calluses induced from different sugarcane varieties is the first negative factor inducing the low efficiency of transformation. Exogenous auxin and cytokinin are necessary to induce cell differentiation and proliferation in in vitro cell culture, such as the embryogenic callus induction of maize[25]. Thus, media for the embryogenic callus induction of different cultivated sugarcane varieties need different exogenous auxin and cytokinin concentrations. For example, high-quality embryogenic calluses of the cultivated sugarcane varieties ROC22 can be obtained by alternately adding high and low 2,4-D concentrations and miniscule amounts of 6-BA to the in vitro culture medium. Sugarcane plants collected 3 months after planting and 2 months before harvesting are suggested to be the best original material for callus induction. The tops of shoots containing the immature leaf whorl must be used to initiate callus induction within 24 h after cutting from the plants (Fig. 1a). First, an immature leaf whorl is transected into thin sections then cultivated in medium with a high concentration (2 mg/L) of 2,4-D; the resulting explant dedifferentiates quickly within 14 d (Fig. 1b). Second, subcultivation is performed in a medium with a low concentration (1 mg/L) of 2,4-D. Tiny embryogenic calluses then form after another 14 d. Third, subcultivation is performed again in a medium containing a low concentration (1 mg/L) of 2,4-D and a very low concentration (0.1 mg/L) of 6-BA. Large and granular embryogenic calluses suitable for transformation form after the subcultivation period (Fig. 1c). The actual concentration of 2,4-D and 6-BA added to the subculture medium for different varieties of sugarcane is based on the status of the calluses growing on the last medium. If callus growth is slow and difficult, the added amount of 2,4-D is increased. If the callus is water-soaked, the concentration of 2,4-D is decreased. Cultivation for callus induction is preferably done in the dark at 28 °C.
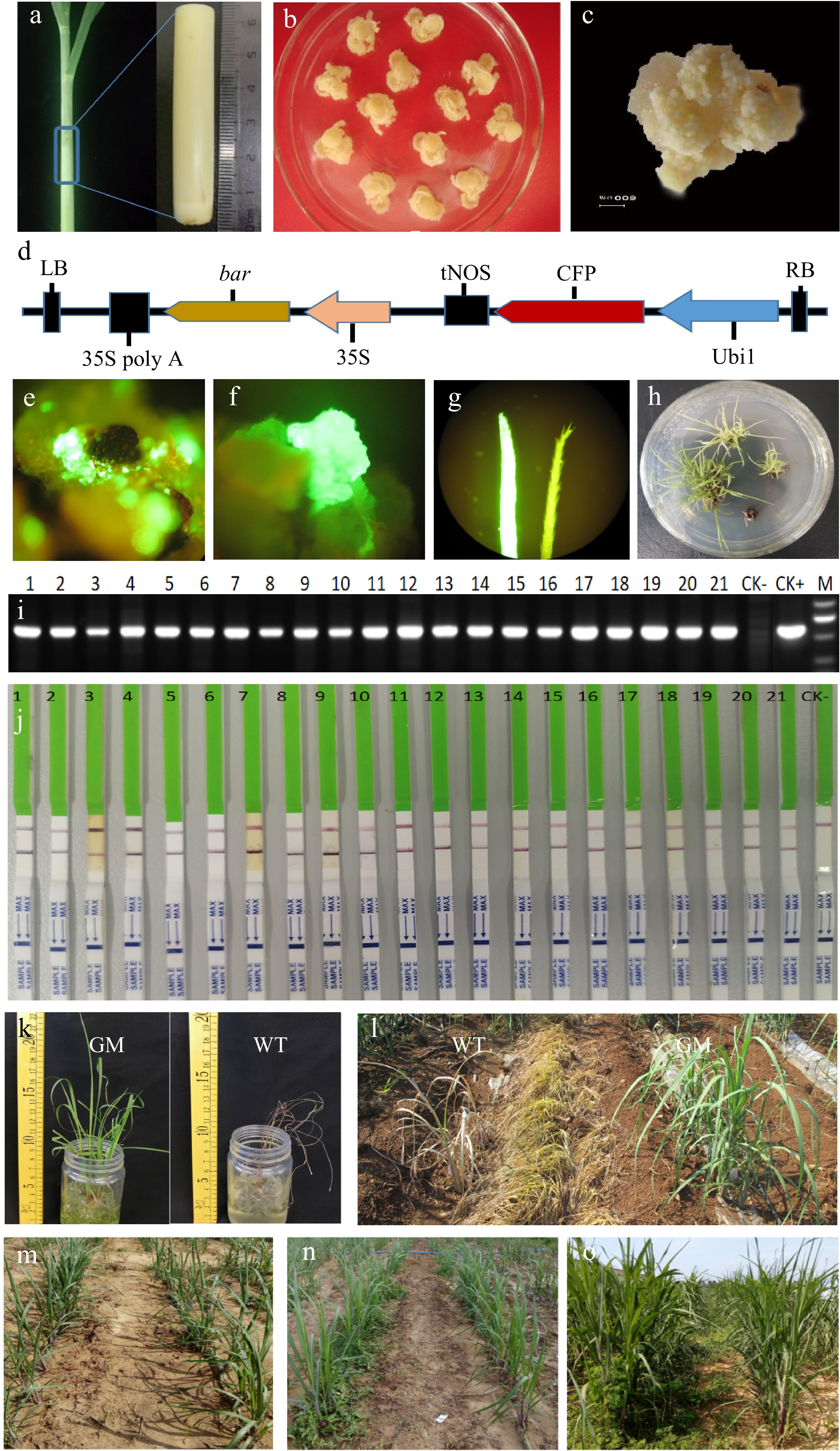
Figure 1.
Efficient sugarcane transgenic system based on herbicide screening. (a) Original explant from sugarcane tillers for callus induction. (b) Two weeks after first induction through the addition of a high concentration (2 mg/L) of 2,4-D to the medium. (c) Two to three weeks after third induction through the addition of a low concentration (1 mg/L) of 2,4-D and a very low concentration (0.1 mg/L) of 6-BA to the medium. (d) Schematic of the pCAMBIA3300-CFP plant expression vector. RB: Right border of pCAMBIA3300; LB: Left border of pCAMBIA3300; Ubi1: maize Ubi 1 promoter; CFP: CFP visible marker gene; tNOS: nopaline synthase terminator; 35S: cauliflower mosaic virus 35S promoter; bar: bar selective marker gene; 35S poly A: cauliflower mosaic virus 35S poly A tail. (e) Fluorescence observation of infected calluses after 7 d of resting cultivation. (f) Fluorescence observation of resistant calluses after 30 d of callus screening. (g) Fluorescence observation of leaves after 14 d of regeneration, Left: leaf of transgenic shoot, Right: leaf of wild-type shoot. (h) Resistant shoots on the rooting medium. (i) Molecular assay for the bar selective marker gene by traditional PCR; CK−: nontransformation shoots; CK+: plant expression vector; and M: DNA marker ladder. (j) PAT/bar protein assay by QuickStix Strips; 1–21: Resistant shoots; CK−: nontransformation shoots. (k) Herbicide tolerance testing of transgenic shoots. GM: transgenic shoots (2.0 mg/mL Basta); WT: Wild-type shoots (0.5 mg/mL Basta). (l) Tenth day after herbicide spraying; GM: transgenic shoots; WT: Wild-type shoots. (m) First day after weed control in the field; Left: Weed control of wild-type shoots by hoeing; Right: Weed control of transgenic shoots by herbicide. (n) Two weeks after weed control in the field; Left: Weed control of wild-type shoots by hoeing; Right: Weed control of transgenic shoots by herbicide. (o) Two months after weed control; Left: Weed control of wild-type shoots by hoeing; Right: Weed control of transgenic shoots by herbicide.
-
Particle bombardment is the main approach used by existing sugarcane transformation studies[26−30]. However, it has several disadvantages, including the low frequencies of positive shoots, high copy numbers of target genes, and instability of the gene construct in the receptor material[31]. Agrobacterium-mediated transformation has advantages over other gene-delivery technologies[32,33]. Our previous research showed that the Agrobacterium strains EHA105 and LBA4404 are suitable for sugarcane genetic transformation. The plant expression vector for vector construction must contain an expression cassette to provide the resistant protein of the selection agent for the positive screening of shoots from transformed calluses. The CaMV 35S promoter has been proven to be sufficiently strong to induce numerous selectable agents, such as the bar, CP-EPSPS, PMI, and NPTⅡ genes, in sugarcane transgenic systems. It can follow at least three other target expression cassettes on the vector aside from the selectable expression cassette, that is, the total integrated fragment could exceed 10 KB between left and right borders. The use of different promoters and terminators for each expression cassette is preferred to avoid vector recombination. The target expression cassettes could be overexpression or RNAi cassettes. Meanwhile, the use of a strong promoter, such as Ubi 1/actin/CaMV 35S, to induce the Cas9 gene and 2–3 u6 promoters to activate 2–3 sgRNA, is preferred for sugarcane editing strategies, such as the CRISPR/Cas9 system. In our previous research, the Agrobacterium strain EHA105 harboring the pCAMBIA3300 plant expression vector containing two expression cassettes was used for transformation to establish our efficient sugarcane transgenic system. One of the expression cassettes was a bar selective marker gene promoted by the CaMV 35S promoter and added with a CaMV 35S polyA tail at the 3ʹ site. The other expression cassette was the CFP visible marker gene promoted by the Ubi1 promoter and terminated with the tNOS terminator (Fig. 1d). The simple vector harboring the visible marker gene is helpful for establishing our efficient transgenic system.
-
A precise transformation protocol increases the infectivity of bacteria and causes receptor explants to absorb the infectious bacterial mixture sufficiently. This situation could increase the efficiency of genetic transformation significantly. Acetosyringone (AS) is a phenolic compound and can activate the vir gene in the plant expression vector, thus increasing the infectiousness of bacteria and the rate of transformation in monocots[34−37]. Hence, it must be added steadily at a suitable working concentration throughout bacterial cultivation. Sonication and vacuum treatment could induce receptor explants to absorb the infectious bacterial mixture sufficiently and enhance Agrobacterium-mediated transformation efficiency[38]. Herein, the sugarcane genetic transformation protocol could be established by combining influencing factors. The precise transformation protocol of our lab is as follows: The Agrobacterium strain harboring the target plant expression vector was streaked on YEP medium containing the appropriate antibiotics and 100 µM AS and grown at 28 °C for 3 d. Then, a single colony was selected and recultured overnight in liquid YEP medium containing the appropriate antibiotics and 100 µM AS at 28 °C. Subsequently, bacteria were collected after centrifugation, resuspended in a starter culture (1/5 strength MS medium + 30 g/L sucrose + 30 g/L glucose + 100 µM AS) and vortexed at 90–100 rpm for 2 h at 28 °C in the dark. Then, the bacterial mixture was diluted to an optical density of approximately 0.3–0.6 at 600 nm. A high density of the bacterial mixture may enhance transformation efficiency but also may induce the high copy number integration of the target genes. A suitable amount of embryogenic calluses (3–5 g) was collected and air dried on a clean bench. Then, the air-dried embryogenic calluses were transferred to an Erlenmeyer flask, added with approximately 50 mL of the bacterial mixture, and shaken slowly at 90–100 rpm for 10 min at 28 °C in the dark. In addition, the embryogenic calluses and bacterial mixture were sonicated (180 W) for 2 min in an ultrasonic cleaner. Then, the bacterial mixture was pipetted out, and 50 mL of fresh bacterial mixture was added again. Subsequently, the embryogenic calluses and bacterial mixture were vacuumed (−0.08 MPa) for 5 min then shaken slowly at 90–00 rpm for another 10 min at 28 °C in the dark. Afterward, the bacterial mixture was pipetted out, and the embryogenic calluses were blotted dry to remove excess Agrobacterium suspension and air-dried for approximately 30 min on a clean bench by using filter paper. Next, the embryogenic calluses were transferred to a Petri dish, sealed with parafilm, then incubated for 3 d at 21 °C in the dark. All of the infected embryogenic calluses were transferred to a resting medium without selection stress and cultured for 7 d at 28 °C in the dark. Subsequently, all embryogenic calluses were transferred to a selection medium containing 2 mg/L Basta (glufosinate ammonium) and cultured for 30 d at 28 °C in the dark. All selected calluses were transferred to a regeneration medium and cultured for 14 d (30 °C and 14 h of light and 28 °C and 10 h of darkness daily). After regeneration, green buds were transferred to a rooting medium and cultured for 30 d under the same conditions. Meanwhile, CFP expression was observed after resting cultivation (Fig. 1e), selection cultivation (Fig. 1f), regeneration cultivation (Fig. 1g, h), and rooting cultivation to estimate transformation efficiency. After rooting cultivation, resistant shoots were sampled for molecular assays via traditional PCR detection (Fig. 1i) and PAT/bar protein detection by using QuickStix Strips (Fig. 1j). The transformation results presented in Table 1 show that on average, 11 transgenic shoots could be obtained from each gram of embryogenic calluses used for transformation. The results of the PCR and QuickStix Strip assays demonstrated that almost 100% of the resistant shoots were positive.
Table 1. Technical service using the efficient transgenic system.
Name of
vector/geneCalluses
used (g)Transgenic shoots provided (lines) Target of genetic transformation Strategy of genetic modification Institutes serviced Date Cry2A 2 17 Pest-resistant genes OE Huazhong Agricultural University 2017.5 Cry1C 2 20 OE 2017.5 Hc-Pro 2 20 Functional gene of yje SCSMV virus OE Yangzhou University 2019.12 ScD27 2 20 Tiller-associated genes OE Sugarcane Research Institute, Yunnan Academy of Agriculture Science 2018.5 ScD10 2 25 RNAi 2019.6 INV 2 15 Sucrose invertase gene OE Institute of Nanfan & Seed Industry, Guangdong Academy of Science 2021.10 FUG 2 15 Haploidy induction gene GE 2019.12 SsWRKY1- OE 2 15 Drought resistance-associated genes OE Yunnan Agricultural University 2020.8 SsWRKY1-RNAi 2 20 RNAi 2020.8 DREB 2 12 Drought resistance-associated genes OE South Subtropical Crops Research Institute, Chinese Academy of Tropical Agriculture Science 2020.9 REMO 2 14 OE 2020.9 MYB8i 2 15 MYB transcription factors RNAi Fujian Agriculture and Forestry University 2020.11 MYB11i 2 12 RNAi 2020.11 ERF99 2 13 Ethylene-responsive factors OE 2020.11 Z6 3 45 JAZ transcription factors OE 2021.9 Z10 3 50 OE 2021.9 VSR 3 80 Vacuolar sorting receptors OE Yulin Normal University 2022.6 R1 3 35 Plant activator polypeptide receptor OE 2022.10 RK1 3 30 Ratoon stunting disease-responsive factors RNAi Sugarcane Research Institute, Guangxi Academy of Agriculture Science 2022.10 * OE: Target gene overexpression; RNAi: Target gene suppression by RNAi; GE: Gene mutation by genomic editing. -
Transgenic shoots screened on the basis of the bar selective marker gene or CP4-EPSPS exhibit herbicide resistance. In our lab, the herbicide-resistant gene bar was used as the selectable marker gene in our transformation system. Resistant shoots were screened by adding herbicide (2 mg/L) to the culture media. The herbicide tolerance of the transgenic shoots was tested in the lab by exposing the shoots to different concentrations of Basta (Fig. 1k). The results showed that all transgenic shoots grew normally even when exposed 2.0 mg/mL Basta. By contrast, the wild-type shoots died when exposed to only 0.5 mg/mL Basta. Thus, the integration of the herbicide-resistant bar gene significantly improved the herbicide resistance of the transgenic shoots, and all of the transgenic shoots obtained by using our transformation system were herbicide-resistant.
-
Herbicide tolerance was also tested in the field. The results showed that on the third day after herbicide spraying (2.0 mg/mL Basta), no significant difference was found between genetically modified and wild-type shoots. However, on the sixth day after herbicide spraying, the leaves of the wild-type shoots were yellowing and shriveled. By contrast, the genetically modified shoots grew healthily in the field. On the tenth day after herbicide spraying, the wild-type shoots died entirely, whereas the genetically modified shoots grew normally throughout the testing period (Fig. 1l). Therefore, the result of field testing coincided with that of herbicide tolerance testing in the lab.
-
Sugarcane transgenic shoots planted in the field could be weed-controlled by using herbicide (200 g/L glufosinate ammonium, YONON Biosciences Co., Ltd., Zhejiang, China), whereas wild-type shoots planted in the field need to be weed-controlled by hoeing (Fig. 1m). The results showed that the weed control of wild-type plants by hoeing was unsustainable. Weeds regrew rapidly 2 weeks after being controlled by hoeing (Fig. 1n) and grew tall and surrounded cane plantlets heavily 2 months later (Fig. 1o). Meanwhile, transgenic shoots were weed-controlled sustainably by using herbicide. Therefore, weed control by herbicide saves labor and costs.
-
Most of the sugarcane breeding institutes of China could not produce any positive transgenic sugarcane shoots, and a few institutes could produce some positive transgenic sugarcane shoots albeit at very low frequencies. After we successfully established the efficient sugarcane genetic transformation system in our lab, we produced numerous transgenic shoots and provided them to almost all of the sugarcane breeding institutes in China through technical services (Table 1). Hence, our system provides a great contribution to molecular breeding and molecular biology research in China. Some gene delivery approaches aim at the character improvement of cultivated varieties, and most aim at the functional study of endogenous and exogenous genes.
-
The bar/Basta selection system was established in our laboratory by using the Agrobacterium-mediated genetic transformation method and a precise transformation protocol. It was proven to be an efficient genetic transformation system that enhanced the quality of induced sugarcane embryogenic calluses. Statistical analysis revealed that 10 or more transgenic shoots could be obtained from each gram of transformed embryogenic calluses used for transformation. In addition, resistant shoots 10 cm in height were obtained approximately 4 months from the initiation of the transformation. Screening revealed that the resistant shoots were almost 100% positive in the molecular assay. All transgenic shoots produced by our transformation system were herbicide-resistant and could be weed-controlled in field trials by using Basta (glufosinate ammonium) herbicide. We are working on the PMI/Mannose[20] and CP4-EPSPS/Roundup (unpublished) systems in our lab in addition to the efficient bar/Basta selection system. Transgenic shoots screened by the CP4-EPSPS/Roundup system are tolerant to Roundup (41% glyphosate), which is cheaper than Basta (20% glufosinate). Thus, the CP4-EPSPS/Roundup system has broad application prospects. By contrast, the released GM sugarcane lines from Brazil, namely, CTB141175/01-A, CTC91087-6, and CTC93209-4, were screened by the NPTII/ G418 and bar/Basta selection systems, and those from Indonesia, namely, NXI-1T, NXI-4T, and NXI-6T, were screened by using the NPTII/G418 and hpt/hygromycin-B selection systems. Therefore, our sugarcane transgenic system is the most advanced in the field. The other results (unpublished) of our research group also showed that our efficient sugarcane transgenic system was effective for ROC22, LC05-136, and GT42, which are the top three cultivated varieties in China. Our system also worked for S. spontaneum, which is an original parent of sugarcane. Thus, it may be effective for all sugarcane germplasms if high-quality embryogenic calluses could be induced. Numerous genome-edited sugarcane lines have also been created by combining genome-editing elements in plant expression vectors and delivering them to the genome by using our genetic transformation system. We produced numerous transgenic shoots of different research targets by using our transformation system and provided them to almost all sugarcane breeding institutes in China through technical services. Therefore, the establishment of our efficient sugarcane genetic transformation system has made a great contribution to the biological study and breeding of sugarcane in China.
This work was supported by Hainan Yazhou Bay Seed Lab (JBGS+B21HJ0302), National Key Research and Development Program of China (2018YFD1000503) and Chinese Agriculture Research System (CARS-170301).
-
The authors declare that they have no conflict of interest.
-
Received 6 December 2022; Accepted 9 June 2023; Published online 19 July 2023
- Copyright: © 2023 by the author(s). Published by Maximum Academic Press on behalf of Hainan University. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Wang W, Wang J, Feng C, Zhao T, Shen L, et al. 2023. Establishment of an efficient transgenic selection system and its utilization in Saccharum officinarum. Tropical Plants 2:11 doi: 10.48130/TP-2023-0011
Establishment of an efficient transgenic selection system and its utilization in Saccharum officinarum
- Received: 06 December 2022
- Accepted: 09 June 2023
- Published online: 19 July 2023
Abstract: Transgenic strategy plays an important role in the biological study and breeding of sugarcane. However, the efficiency of sugarcane transgenic systems remains disappointing to breeders. Various cultivated varieties are recalcitrant to genetic transformation, and only a few sugarcane research institutes could successfully obtain positive transgenic lines. In our previous research, three kinds of sugarcane transgenic selection systems, namely, the PMI/Mannose, CP4-EPSPS/glyphosate, and bar/Basta selection systems, were successfully established. Among these systems, the bar/Basta selection system was the most efficient. By applying this selection system, 10 or more transgenic shoots could be obtained from a gram of embryogenic calluses. In addition, the resistant shoots obtained after screening were almost 100% positive for the molecular assay, and all of the transgenic shoots showed high herbicide tolerance in lab tests and field trials. Herein, the key points/steps, advantage and contribution to sugarcane studies and breeding in China of the efficient bar/Basta sugarcane transformation system are presented and discussed.
-
Key words:
- Sugarcane /
- Genetically modified /
- Screening system /
- Agrobacterium-mediated /
- Herbicide tolerance /
- bar gene












