-
One of the most evident consequences of global warming is the escalation of extreme weather events, which enables abiotic stress to occur more frequently[1,2]. Vegetable crops are diverse in nature including cucumber, watermelon, tomato, pepper, and lettuce, etc. All have significant economic, nutritional, and aesthetic value. Adverse light and temperature are two major environmental factors that can trigger a variety of common abiotic stressors including low light, high light, cold, and heat stresses, etc. These stressors will not only damage vegetable crops, but will also have a substantial impact on their development, yield, and quality[3].
In plants, alternative splicing (AS) is disproportionately used to produce critical specific mRNA splicing variants by the spliceosome for coping with fluctuating light and temperature environments. The spliceosome is a dynamical complex composed of hundreds of splicing factor (SF) proteins responsible for recognizing cis-elements, removing introns, and selectively ligating exons of pre-mRNA[4−6]. Almost 85% of genes are multiexon, and 70% of them are alternatively spliced in Arabidopsis[7]. In recent times, numerous studies have been carried out to determine the role of AS in various plant species. Splicing variants of SL (strigolactone) receptor gene D14 and REGULATOR OF LEAF INCLINATION 1 (RLI1) act as very critical roles in controlling key agronomic traits in rice[8−10]. Furthermore, the AS of heat-shock transcription factors (HSFs), hy5 homolog (HYH), and phytochrome interacting factors (PIFs) from Arabidopsis are regulated, and these induced splicing variants play significant roles under diverse light and temperature conditions[5,11−13]. So, AS is an important and widely used strategy for plants to cope with light and temperature stresses.
SFs, including glycine-rich RNA-binding proteins (GRPs), serine/arginine-rich (SR) proteins, and small nuclear ribonucleoprotein particles (snRNPs), are essential components of the spliceosome that govern AS[14,15]. The majority of these SFs have been recognized as crucial for stress tolerance, and mutants of these SFs are usually defective in growth and development, with poor tolerance against light and temperature stresses[16−18]. Hence, studies on AS events and SFs involved in the cascade of light and temperature stress signals is emerging and crucial. In this review, we first summarize the current understanding of the core spliceosome and discuss the functions of different types of AS events and SFs in plants. Then, we briefly discuss the classical regulatory pathways in the control of AS under adverse light and temperature conditions. Moreover, knocking out the gene by CRISPR/Cas9 technique or overexpressing its major coding splicing variant usually will lead to undesired phenotype changes in plant growth and development. Thus, a possible strategy is proposed for discussion. Through CRISPR/Cas9 technology we can editing the splicing codes or splicing factors that can manipulate the AS to produce splicing variants with distinct function for understanding the mechanism of AS and developing genetic improvements in vegetable crops.
-
In eukaryotes, most genes are interrupted by introns and protein-coding sequences known as exons. gDNA is transcribed by RNA polymerase-II (Pol-II) to produce pre-mRNA with introns cotranscriptionally[19]. Pre-mRNAs contain critical cis-elements, including splice site (SS) consensus sequences (5'SS, 3'SS, branch point, and polypyrimidine tract), exonic splicing enhancers/silencers (ESEs/ESSs), and intronic splicing enhancers/silencers (ISEs/ISSs) (Fig.1). The cis-acting components listed above are referred to as splicing code, and they all have a substantial impact on splice-site selection and AS consequences[20,21]. First, the recognized efficiency of splicing factors is based on how intense the SS consensus sequences are[22]. If the SSs or splicing code sequences are altered, their relative strengths will change. This might end up causing the spliceosome to recognize new SSs and change how pre-mRNA has been spliced[23]. Secondly, flanking pre-mRNA regulatory sequences (ESEs/ESSs and ISEs/ISSs) served as the binding site for different splicing factors[20]. Among them, SR proteins are responsible for binding ESEs and ISEs whereas heterogeneous nuclear ribonucleoproteins (hnRNPs) mainly bind ESSs and ISSs. Binding the flanking pre-mRNA regulatory sequences by these SFs can help snRNPs choose the right SS. Meanwhile, these different types of SFs and cis-acting elements show antagonistic functions in selecting splice sites[22].
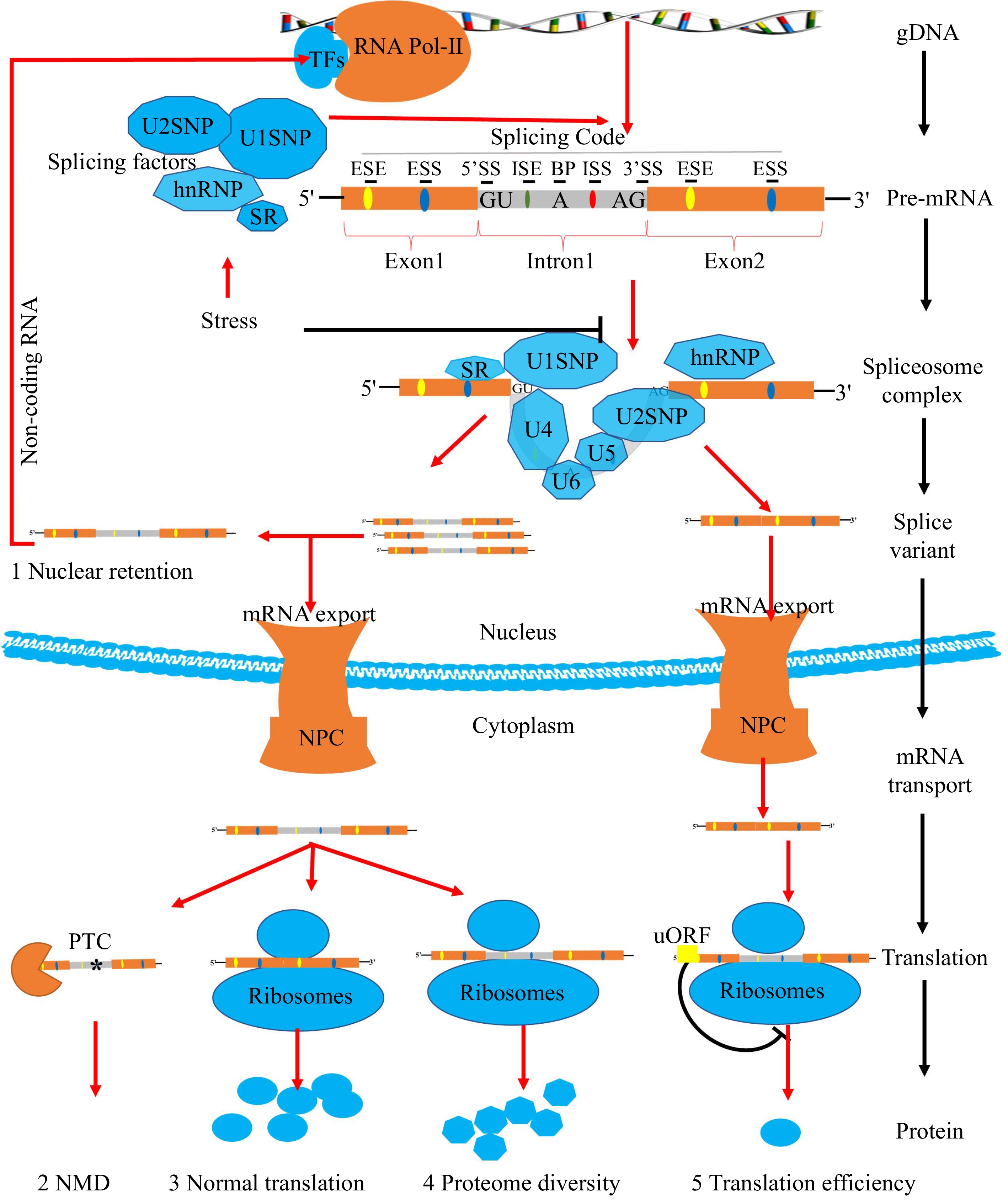
Figure 1.
Pre-mRNA splicing process of plants. In plants, most coding sequences of genes are interrupted by introns. Transcription factors (TFs) recognize promoters and guide the expression of genes. The gDNA is transcribed by RNA polymerase-II (Pol-II) to produce pre-mRNA with both exons and introns. Pre-mRNA owns critical splicing codes, including 5'SS, 3'SS, branch point, polypyrimidine tract, ESEs/ESSs, and ISEs/ISSs. SFs are responsible for recognizing splicing codes. The active spliceosome removes introns, ligating exons of pre-mRNA selectively to produce splicing variants. Stresses have significant effects on AS through SFs. Different splicing variants will have multiple molecular consequences. Firstly, the nuclear pore complex (NPC) will prevent certain intron-containing mRNAs leakage into the cytoplasm and leading to nuclear retention events. Secondly, mRNA transcripts containing premature translation-termination codons (PTCs) with 3'-untranslated regions (3'UTRs) (≥ 350 nt) can be degraded by nonsense-mediated RNA decay (NMD). Finally, the remaining mRNA transcripts will be translated into different spliced proteins by ribosome with different translation efficiency due to the presence of (upstream open reading frames) uORFs.
Numerous splicing factors recognize distinct splicing codes and can assemble a highly dynamic spliceosome complex composed of five core uridine-rich snRNPs (U1, U2, U4, U5, and U6), hundreds of small nuclear RNA (snRNA) genes, and accessory proteins[22]. Sequence comparison and motif searches in Arabidopsis identified 395 splicing-related proteins encoding genes[14]. Splicing-related proteins are referred to as splicing factors in this context. Most of them are different types of RNA-binding proteins responsible for recognizing the splicing code on pre-mRNA[21]. For example, pre-mRNA-processing factor 8 (Prp8) was a scaffolding protein of the U5 snRNP that regulates 5'SS and 3'SS utilization. It shares a multidomain structure characterized by one RNA recognition motif (RRMs), which bind to the pre-mRNA targets, and two U6/5-snRNA binding sites involved in snRNA–protein interactions[24]. More and more splicing variants and SFs have been discovered as crucial for various stress responses in Arabidopsis and rice, but vegetable crops have achieved modest progress in this field.
The modes of alternative splicing are complicated and diverse, including exon skipping (ES), intron retention (IR), etc. (Fig. 2a). Among them, IR is the most prevalent. More than 51% of genes producing AS transcripts show IR events in Arabidopsis, but in metazoans, ES is prevalent[25,26]. Earlier studies revealed that the majority of splicing variants induced by stress were ineffective[27] . Yet, increasing evidence suggests that mRNA splicing variants perform active functions in response to stressful conditions. At first, the nuclear pore complex (NPC) will prevent intron-containing splicing variants from leakage into the cytoplasm[28] . Usually, these splicing variants in the nucleus might act as the non-coding RNA rather than coding proteins. The activating signal cointegrator complex 3 (ASCC3) gene, for example, can be spliced into coding and non-coding transcript isoforms with antagonistic activities in regulating transcription recovery following UV-induced DNA damage[29]. Secondly, splicing variants containing premature translation-termination codons (PTCs) with 3'UTRs (≥ 350 nt) can be degraded by nonsense-mediated RNA decay (NMD)[30]. Then, the splicing variants in the cytoplasm will be translated into different spliced proteins (Fig. 2b). Furthermore, if AS occurs in 5'UTR, it will not affect the coding sequence but change the presence of uORF (Fig.1 and Fig. 2b). uORF is widely present in the 5'leader sequences of 30–50% of eukaryotic mRNAs. The translation of uORF can greatly repress the translation efficiency of the downstream major coding sequences[31].
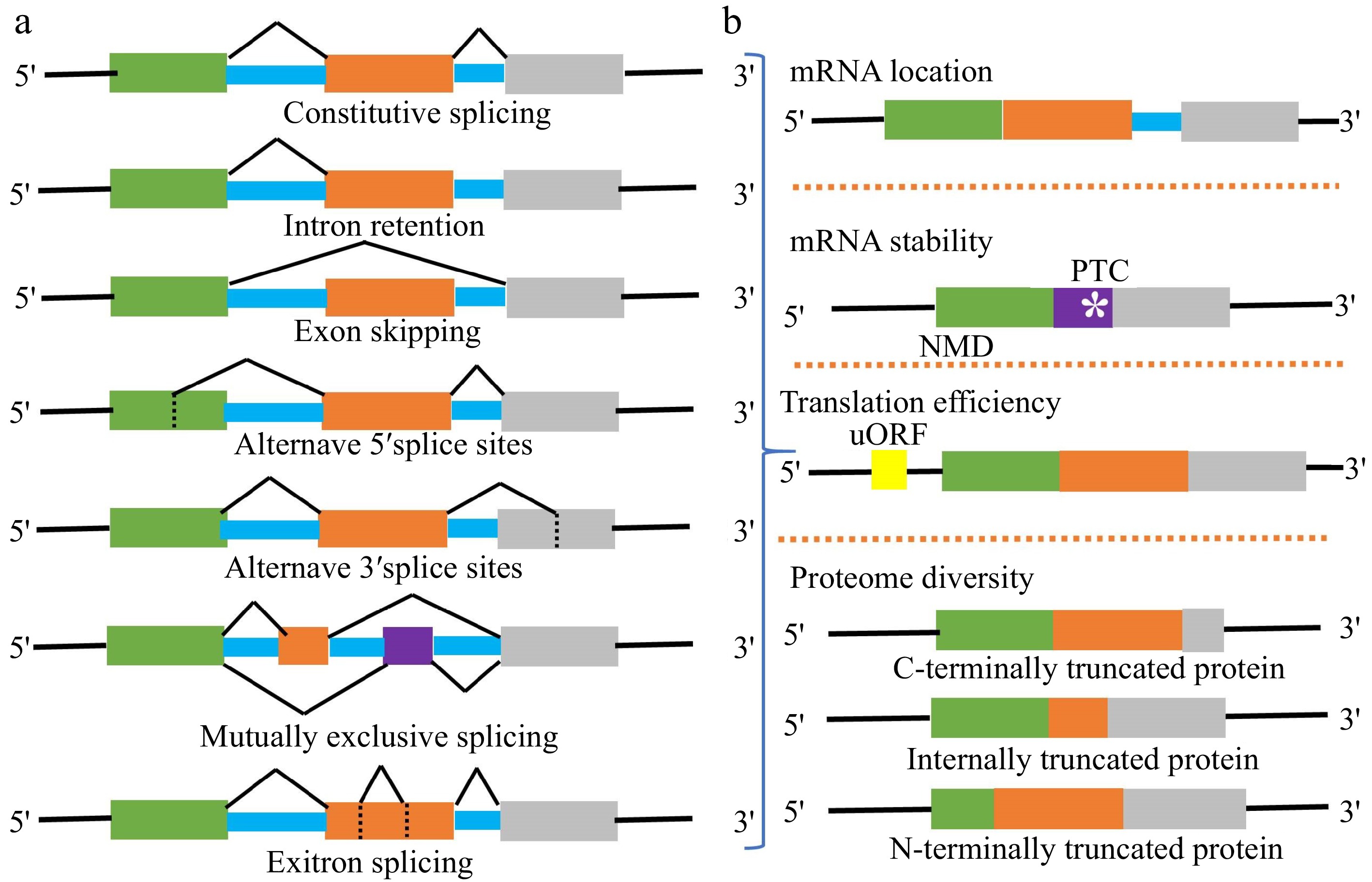
Figure 2.
(a) Major types of AS events in plants include constitutive splicing (CS), intron retention (IR), exon skipping (ES), alternative 5' splice sites (A5SS), alternative 3' splice sites (A3SS), mutually exclusive splicing (MES), and exitron splicing (EIS). Usually, an mRNA splicing variant is formed by one or more AS events. (b) Different AS event combinations lead to generate multiple splicing variants from one gene locus. The subcellular location, stability, translation efficiency, and coding sequence of splicing variants are different.
-
Light and temperature are the most important environmental factors regulating plant growth and development. Key factors in the light signaling pathway, including PIF3, PIF6, ELONGATED HYPOCOTYL5 (HY5), suppressor of phyA-105 3(SPA3), and cryptochromes 1 (CRY1), are proven to suffer AS[5]. PIF3 was extensively investigated among them, and red light greatly increased amounts of the intron retained in PIF3 5'UTR. A uORF contained within this intron caused an 80% reduction in PIF3 protein levels[32]. In addition to PIF3, AS pattern change of PIF6 contributes to seed dormancy response[33,34], and the selection of alternative 5’SS within SPA3 intron 4 leads to increased truncated nonfunctional SPA3 protein[12]. Interestingly, phytochromes (PHYB and PHYA) are responsible for light-dependent AS of SPA3, PIF3, and PIF6[12,35]. The roles of these phytochromes in AS will be discussed later.
Heat-shock transcription factors (HSFs) are vital for plant heat stress tolerance. HsfA2, a key regulator in response to heat stress in Arabidopsis, is also heat stress-induced spliced (a cryptic splice site in the intron) to produce a small truncated HsfA2 isoform with important function under heat stress conditions[36,37]. Another type of transcription factor bZIP60 involved in heat stress tolerance by controlling unfolded protein response[38−42] , and its mRNA can be alternatively spliced by inositol requiring enzyme1 (IRE1) to produce two transcripts encoding different functional spliced proteins[43]. Moreover, flower time and fertility could also be greatly affected by temperature variation. FLOWERING LOCUS M (FLM) splicing variants play key roles in flower time. The FLM-β is predominately formed at low temperatures and prevents precocious flowering. By contrast, the FLM-δ encodes a truncated protein with impaired DNA binding and acts as a dominant-negative activator of flowering at higher temperatures[44]. Temperature-sensitive genic male sterility (TGMS) line is important for the breeding of hybrid crops. The splicing of CALLOSE SYNTHASE5 (CalS5) pre-mRNA is involved in controlling TGMS[45,46]. So, light and temperature change significantly affect AS patterns, which are essential for plant stress adaption.
-
Light dramatically affects alternative splicing[6,47]. Critical genes of light signaling pathways in plants, including PHYB, PHYA, CRY2, and CONSTITUTIVELY PHOTOMORPHOGENIC 1 (COP1), act similarly to SFs in determining AS (Fig. 3 & Table 1). Among them, PHYB regulates photomorphogenesis, thermomorphogenesis, and freezing tolerance[48,49]. PHYB could also directly interact with SPLICING FACTOR FOR PHYTOCHROME SIGNALING (SFPS) and red-light responses in CRY1CRY2 background1 (RRC1) in nuclear speckles to synergistically regulate pre-mRNA splicing of a group of genes involved in light signaling and circadian clock pathways[12,35,50−52]. Furthermore, PHYA regulates AS via interaction with the Arabidopsis homologue of human CNOT9 (NOT9B)[53]. During photomorphogenesis, COP1 can regulate AS of U2 small nuclear ribonucleoprotein auxiliary factor 65A (U2AF65A) and SR30 by interacting with U2AF65-associated protein (UAP56)[54]. Hence, we can conclude that these proteins in light signaling pathways can directly regulate SFs in order to modulate AS in response to changes in the light environment.
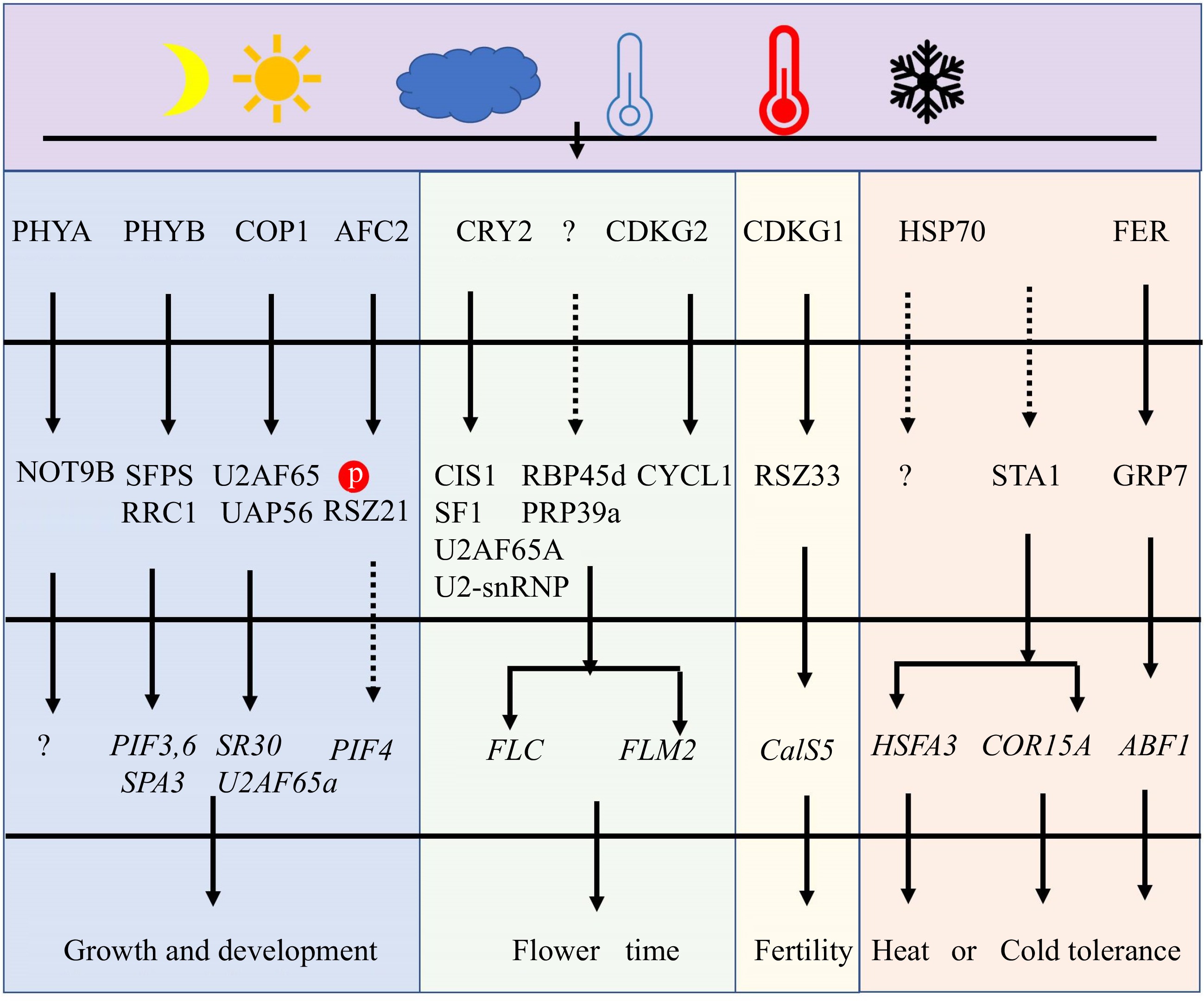
Figure 3.
The major pathway for regulating pre-mRNA AS under light and temperature stresses. At first, some SFs could act as sensors to perceive the changes in light and temperatures. Secondly, those sensors will phosphorylate or interact with other SFs and change their location and activity. These modifications of SFs will affect AS by controlling spliceosome function. Finally, plant growth, development, flower, fertility, and tolerance are regulated under adverse light and temperature conditions.
Table 1. Splicing factors involved in light and temperature stress adaption.
Annotation Name Species Gene ID Function Reference DEAD-box RNA heatlicases RH42 Rice Os08G0159900 Cold Tolerance Lu et al.[65] U1 snRNP Prp39a Arabidopsis AT1G04080 Temperature-induced flowering time Chang et al.[73] hnRNPs RBP45d Arabidopsis AT5G19350 Temperature-induced flowering time Chang et al.[73] Phytochrome CRY2 Arabidopsis AT1G04400 Blue-light regulation of thermosensory flowering Zhao et al.[74] mRNA binding SF1/BBP Arabidopsis AT5G51300 Temperature-induced flowering time Lee et al.[71] hnRNP A/B family UBA2c Arabidopsis AT3G15010 Light and temperature-induced flowering time Zhao et al.[72] Core proteins (Sm) SmE-b/PCP/SME1 Arabidopsis AT2G18740 Arrest of growth and male sterile at
low temperatureCapovilla et al.[17] U1 snRNP Luc7a Arabidopsis AT3G03340 Low temperature resulted in arrest
of growthAmorim et al.[63] U1 snRNP Luc7b Arabidopsis AT5G17440 Low temperature resulted in arrest
of growthAmorim et al.[63] U1 snRNP Luc7-rl Arabidopsis AT5G51410 Low temperature resulted in arrest
of growthAmorim et al.[63] Glycine rich protein GRPs-7 Arabidopsis AT2G30560 Cold tolerance Kim et al.[62] DEAD box RNA helicase Prp5-1b/RCF1 Arabidopsis AT1G20920 Cold tolerance and cold acclimion Guan et al.[59] U5 snRNP U5-200-2b/Brr2b Arabidopsis AT2G42270 Cold tolerance and cold acclimion Guan et al.[59] DEAD-box RNA helicase U2B"a/U2B"-LIKE Arabidopsis AT1G06960 Cold tolerance and cold acclimion Calixto et al.[64] SR protein kinase AFC3/AME3 Arabidopsis AT4G32660 Cold tolerance and cold acclimion L. Savitch, unpublisheatd data 17S U2 snRNP GEMIN2 Arabidopsis AT1G54380 Cold tolerance and cold acclimion Schlaen et al.[61] U5 snRNP U5-102KD/STA1 Arabidopsis AT4G03430 Cold tolerance and cold acclimion;
heat tolerance and heat acclimionKim et al.[66];
Guan et al.[59]Inositol-requiring enzyme-1 IRE1b Arabidopsis AT5G24360 Heat stress responses Deng et al.[39]; Moreno et al.[38] Inositol-requiring enzyme-1 IRE1a Arabidopsis AT2G17520 Heat stress responses Deng et al.[39]; Moreno et al.[38] hnRNP HOS5/RCF3 Arabidopsis AT5G53060 Heat stress tolerance Guan et al.[60] HSP70-3 HSP70-3 Arabidopsis AT3G09440 Heat stress tolerance Song et al.[56] HSP70-4 HSP70-4 Arabidopsis AT3G12580 Heat stress tolerance Wang et al.[57] HSC70-1 HSC70-1 Arabidopsis AT5G02500 Heat stress tolerance Tiwari et al.[58] Cyclin-dependent kinase CDCL2p110-1 Arabidopsis AT1G67580 High temperature-induced male fertility defects Cavallari et al.[70]; Zheng et al.[69]; Nibau et al.[68] Cyclin-dependent kinase CDCL2p110-2/CDKG1 Arabidopsis AT5G63370 High temperature induce fertility defects Cavallari et al.[70]; Zheng et al.[69]; Nibau et al.[68] SR protein Kinase AFC2 Arabidopsis AT4G24740 Thermosensors and thermomorphogenesis Lin et al.[88] Phytochrome PHYB Arabidopsis AT2G18790 Thermosensors and thermomorphogenesis;
photoreceptors and photomorphogenesisJung et al.[48]; Shikata et al.[12,51]; Hartmann et al.[52]; Xin et al.[35,50] Phytochrome PHYA Arabidopsis AT1G09570 Photomorphogenesis Shikata et al.[12]; Schwenk et al.[53] E3 ubiquitin ligase COP1 Arabidopsis AT2G32950 Photomorphogenesis Li et al.[54] 17S U2 associated proteins SR140-1/RRC1 Arabidopsis AT5G25060 Photomorphogenesis Shikata et al.[12,51]; Hartmann et al.[52]; Xin et al.[35,50] SFs are also discovered to play a critical role in regulating plant temperature stress tolerance. Heat shock proteins (HSP70s), as a spliceosome component, are involved in the repair of AS during heat stress[55]. In Arabidopsis, HSP70-3, HSP70-4, and heat shock cognate protein 70–1 (HSC70–1) are all required for heat stress tolerance[56−58]. Apart from heat shock protein, a nuclear-localized SF with a K homology (KH) domain, REGULATOR OF CBF GENE EXPRESSION 3 (RCF3) serves as a negative regulator in heat stress tolerance[59,60]. For cold stress, SFs including RCF1, U5 small nuclear ribonucleoprotein helicase (Brr2b)[59],Gemin2[61], GRP7[62], the LAMMER kinase 3 (AME3), LETHAL UNLESS CBC 7 (LUC7)[63], and U2 small nuclear ribonucleoprotein particle (snRNP)-specific proteins (U2B”-LIKE)[64] play important roles in cold acclimation and tolerance in Arabidopsis. In rice, DEAD-box RNA Helicase42 (OsRH42) modulates pre-mRNA splicing at low temperatures by directly interacting with U2 small nuclear RNA. OsRH42-knockdown and OsRH42 overexpression dramatically affect the pre-mRNA splicing pattern, results in retarded plants growth and impaired cold tolerance[65]. Interestingly, the U5-snRNP-interacting protein homolog STABILIZED1 (STA1) is necessary for both cold and heat stress tolerance (Fig. 3 & Table 1). STA1 acts as a high temperature-induced SF for controlling AS of HSFA3[66]. Cold stress has also been shown to enhance STA1 gene expression, and the sta1-1 mutant is defective in splicing the cold-induce gene COR15[67]. Thus, the preceding studies indicate that the majority of SFs positively influence temperature stress tolerance.
Under adverse temperature conditions, SFs can also have a significant impact on plant development, flowering, and fertility. Sm protein E1 (SME1), for instance, is a temperature-specific SF in Arabidopsis, and mis-splicing WUSCHEL and CLAVATA3 rendered sme1 mutants growth and male sterility[17]. Similarly, CDKG1 regulates fertility under temperature stress and can interact with splicing factor Arginine/serine-rich zinc knuckle-containing protein33 (RSZ33), U2AF65A, and RS-containing zinc finger protein22 (SRZ22) to facilitate the splicing of the sixth intron of CalS5, consequently modifying pollen wall development (Fig. 3)[68,69]. CDKG1 can also be spliced into two transcript variants (CDKG1L and CDKG1S), and the ratio between long and short forms is regulated by temperature and CDKG2[46,70]. Furthermore, SFs are primarily engaged in flowering time control. In response to temperature stress, AtSF1[71], UBIQUITIN-SPECIFIC PROTEASE 1-ASSOCIATED PROTEIN 2C (UBA2c)[72], RNA binding protein 45d (RBP45d)[73], CRY2[74], U2AF65A[74], CRY2 INTERACTING SPLICING FACTOR 1 (CIS1)[74], PRP39a, CYCLYN L1 (CYCL1), and CDKG2[68], all of them can govern the AS of FLM to fine-tune flowering time. Among them, CDKG2 together with its cognate CYCL1 controls the AS of FLM. CRY2 can promote CIS1 binding to pre-mRNA of FLM, then CIS1 interact with SF1 and U2AF65A to recruit U2-snRNP to control AS of FLM. Thus, the CRY2–CIS1–U2AF65A/SF1–FLM signaling pathway shows the specific roles of AS under light and temperature stresses (Fig. 3).
-
The development of essential vegetable traits including yield, shape, color, nutritional value, flower, and fruit, is usually dependent on strict temperature and light requirements. Adverse light and temperature will produce low-quality vegetable crops, drastically reducing their economic and nutritional value[75]. For instance, low light and high temperature stress will lead to the formation of slender seedlings with a long stem, smaller leaf, and short root. Thus, these seedlings typically have lower stress tolerance, are susceptible to lodging, and difficult to transplant (Fig. 4a). In order to ensure a stable supply of vegetables and fruits during the off-season, protected fruit trees and vegetables are generally cultivated in early spring and late autumn. But still, low temperatures and low light stress are prevalent during this season, resulting in significant reductions in agricultural yields and quality. Temperature stress, in particular for fruit development, can considerably alter the accumulation level of fruit sugar content, and the quality and intensity of light have a substantial impact on the color formation[76−78].
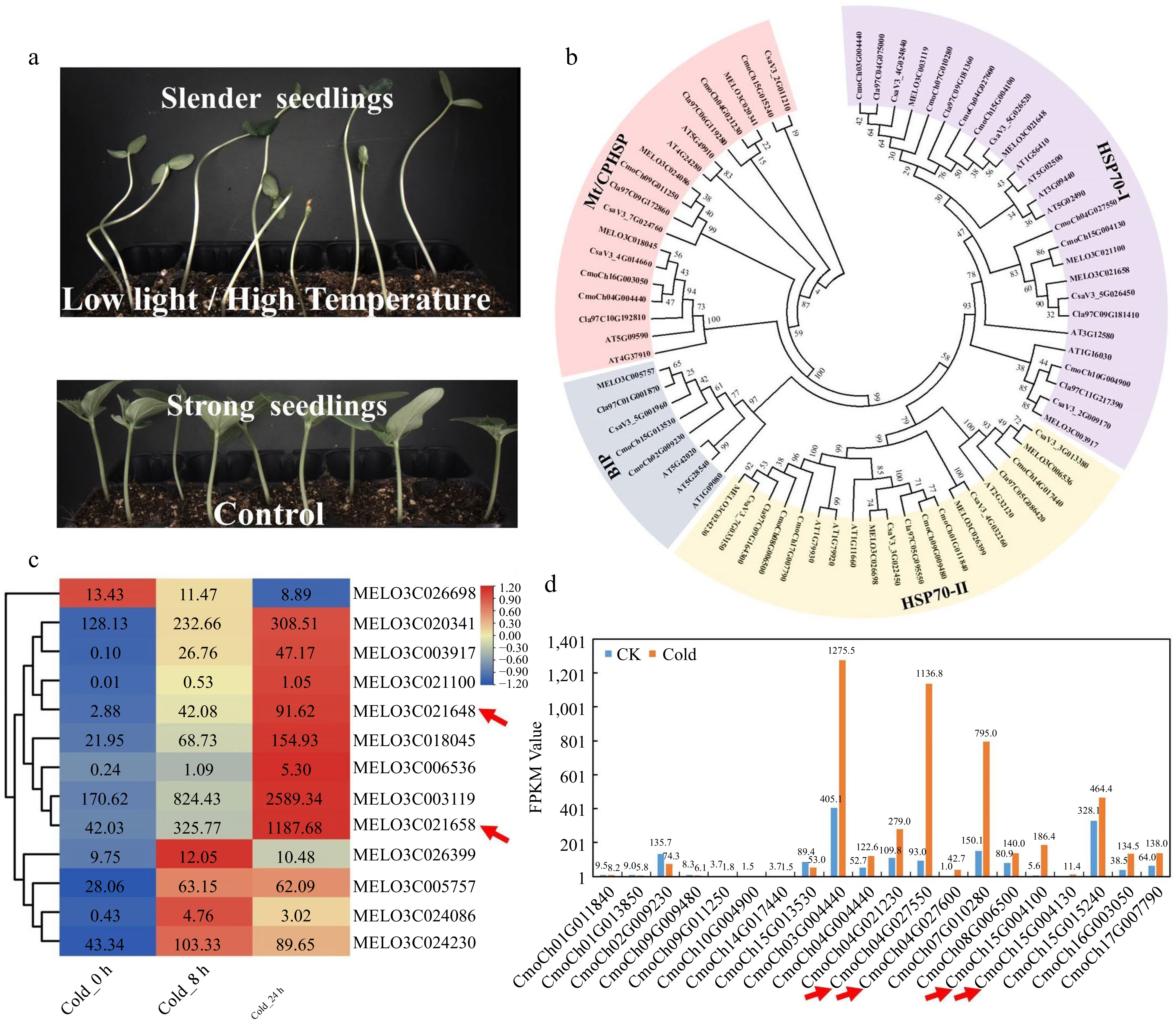
Figure 4.
Expression patterns analysis of the HSP70 gene family members in cucurbit crops. (a) Low light or high temperature stress causes slender cucumber seedlings in the breeding greenhouse. (b) The phylogenetic tree of the HSP70 gene family from cucumber, pumpkin, watermelon, tomato, melon, and Arabidopsis. (c) Heatmap of melon HSP70 gene family after 8 h and 24 h cold treatment. (d) Gene expression patterns of the pumpkin HSP70 gene family after 24 h cold treatment.
AS has been shown to be an efficient post-transcriptional regulatory mechanism in Arabidopsis for coping with fluctuating light and temperature stressors[6,7,12,47,79]. However, the distinctive roles of SFs and AS in vegetable crops have not been thoroughly investigated under light and temperature constraints. Vegetable crops have only published a couple of research results associated to AS regulation during light and temperature stress in recent times. In cucumber, AS of CsGA2ox8 is affected by high light to control hypocotyl elongation[80]. In lily, heat stress leads LlHSFA3B intron intention to produce the splicing variant LlHSFA3B-III with PTCs, which could increase plant tolerance of heat stress by encoding a truncated protein[11]. In tomato, it was found that intronic polymorphisms of SlHsfA2 affected the splicing efficiency and heat stress tolerance[81]. The findings demonstrate that AS is also crucial in the response of vegetable crops to light and temperature stressors.
-
In Arabidopsis, 395 protein-coding genes involved in AS are identified and classified into five categories (91 small nuclear ribonucleoproteins, 109 splicing factors, 60 splicing regulation proteins, 84 novel spliceosome proteins, and 51 possible splicing related proteins)[14]. Many splicing factors have been shown to be essential for stress tolerance. However, they have not been identified and analyzed effectively in most horticultural crops. Low light or high temperature stress are usual in the greenhouse when seedlings are being raised. It can cause slender seedlings with long hypocotyl and lower lodging resistance, especially in cucurbit crops (Fig. 4a). To find SFs involved in seedlings, we take vegetable crops (cucumber, pumpkin, watermelon, tomato, melon) as an example to carry out blastp search. One thousand seven hundred and eight SF protein sequences were retrieved and classified, including cucumber 326, melon 304, pumpkin 429, watermelon 303, and tomato 346 (Supplemental Table S1). The following has established a solid platform for us to investigate the role of splicing factors under light and temperature stresses. We identified that temperature stress had a significant effect on the expression of HSP70s by examining the expression patterns of all the SFs genes under light and temperature stresses. Hence, we systematically identified the HSP70 gene family and constructed the phylogenetic tree and heatmap (Fig. 4b). Surprisingly, low temperature stress substantially induced the majority of HSP70s gene family members, with HSP70−4 the most distinctive among them (Fig. 4c & d). HSP70 is a spliceosome component that plays a vital role in the repair of AS during temperature stress[55]. In cucurbit crops, its members are doubled, including two homologs in melon (MELO3C021658 and MELO3C021640), four homologous genes in pumpkin (CmoCh04G027550, CmoCh04G027600, CmoCh15G004100, and CmoCh15G004130), and all of which are highly temperature-responsive (Fig. 4b−d). Unfortunately, their involvement in controlling AS and adaptation to the light and temperature stressors of vegetable crops are unclear and require extensive research.
-
Traditionally, most studies only examine at one major splicing variant, and it is also presumed that the splicing variant of interest can be translated into proteins. One gene can produce several or even dozens of splicing variants through AS. Yet, only some splicing variants can be translated into distinct proteins. Other splicing variants without coding capacities maybe act as active non-coding RNA (Fig. 1). It is not enough to explain the gene function by analyzing one or two splicing variants with coding capacities. In plants, splicing factors and splicing codes are two major intrinsic factors in the control of AS. Many studies have proven that DNA polymorphism in splicing code regions could lead to changes in AS and phenotype[82−86]. SFs are another factor controlling AS, and there is a complex hierarchical regulatory relationship between SFs (Fig. 3). SFs including receptor-like kinase receptor FERONIA (FER) and LAMMER kinase 2(AFC2) could act as sensors to detect changes in suboptimal light and temperature and subsequently phosphorylate or interact with other SFs (GRP7 and RSZ21) to alter their location or activity in regulation of AS of Abscisic acid response element binding factor 1 (ABF1) and PIF4[87−89]. Furthermore, the interaction between SFs and splicing codes cannot be neglected since SFs can also influence splicing fidelity by restricting the usage of novel splicing code[23]. Consequently, a thorough investigation of splicing codes, splicing factors, and their effects on AS under stress will allow us to better understand and exploit it in the future for vegetable crops.
CRISPR-Cas9 is the most advanced genome editing approach, and it has been extensively used in precision breeding of important cereals and vegetable crops[90,91]. AS modulates many thermomorphogenesis and photomorphogenesis genes in Arabidopsis, including CRY1, PHYB, and EARLY FLOWERING 3 (ELF3). They are also important in modulating seedling elongation under low light or high temperature stresses[92]. Yet, knockout of these genes by CRISPR/Cas9 will make plants extremely susceptible to low light and high temperature. They will also cause undesirable alterations in plant growth and development, including flowering timing, fruit set, seed germination, and so on[93]. Whether we can edit their splicing code (intron or UTR region) or splicing factors to manipulate AS of these genes by CRISPR/Cas9 to produce critical splicing variants for inhibiting excessive elongation but without other undesired effects? Thus, we propose an application potential of this AS mechanism for vegetable crops in (Fig. 5) and hope that this research strategy will promote studies on AS regulation in combination with gene editing technology under light and temperature stresses, which can be used in the future for the genetic improvement of vegetable crops.
This research was supported by Guangdong Basic and Applied Basic Research Foundation (2021A1515110515), the Innovation Fund Projects of Guangdong Academy of Agricultural Sciences (202208 and 202149), the Science and Technology Project of Zhaoqing City (2021N004), and the Guidance Project for Young People of Guangdong Academy of Agricultural Sciences (R2020QD-064).
-
The authors declare that they have no conflict of interest.
- Supplemental Table S1 Vegetable crops splicing-related proteins.
- Copyright: © 2023 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Cao H, Wu T, Shi L, Li Y, Zhang C. 2023. Alternative splicing control of light and temperature stress responses and its prospects in vegetable crops. Vegetable Research 3:17 doi: 10.48130/VR-2023-0017
Alternative splicing control of light and temperature stress responses and its prospects in vegetable crops
- Received: 20 December 2022
- Accepted: 17 March 2023
- Published online: 02 June 2023
Abstract: Vegetable crop quality and productivity are impaired by adverse light and temperature stresses, such as low light, cold, and heat stress. Alternative splicing is a powerful strategy for coping with these abiotic stresses by producing multiple mRNA splicing variants with different subcellular locations, translation efficiency, and coding sequences. Pre-mRNA is alternatively spliced by the spliceosome consisting of different splicing-related proteins termed splicing factors. Recent studies have shown that splicing factors and splicing variants have diverse effects on the development, flowering, and fertility of model plants under light and temperature stresses, but there is very little information available for vegetable crops. Therefore, we conducted a comprehensive literature review on the roles of alternative splicing in regulating plants' responses to light and temperature stress. Several suggestions for future studies and implementation in the field of vegetable crops are presented. We can manipulate alternative splicing using CRISPR/Cas9 technology to produce splicing variants with diverse roles for improving performance of vegetable crop under stress environments.
-
Key words:
- Alternative splicing /
- Splicing factor /
- Light and temperature stresses /
- Spliceosome /
- CRISPR/Cas9













