-
R2R3-MYB transcription factors (TFs) are extensively involved in plant development, secondary metabolism, cell differentiation, and stress response[1−4]. A number of studies have found that plant R2R3-MYB TFs show diverse evolutionary patterns, with some R2R3-MYB TFs presenting conservative functions, while others work as lineage-specific controllers[1,5−7]. Because R2R3-MYB TFs have significant genetic and functional diversity, it is of interest to study the evolution of the R2R3-MYB TF family in crops.
The essential biological functions of the R2R3-MYB type protein MYB36 have been investigated in detail in Arabidopsis and rice plants[7−12]. The function of MYB36 has been studied in some important plants, such as cucumber (Cucumis sativus L.), cotton (Gossypium hirsutum), Salvia miltiorrhiza and Salvia castanea. In Arabidopsis, AtMYB36 regulates the development of whole roots[8−10,13]. AtMYB36 regulates the developmental transition of roots from proliferation to differentiation by inhibiting the TFs that regulate cell division and differentiation[9]. During the maturation of the root endodermis, AtMYB36 directly regulates the expression of CASP1, PER64 and ESB1, which are the main genes involved in the formation of the Casparian strip[8]. Moreover, overexpression of AtMYB36 enhanced the deposition of suberin in the root endodermal cell layer, and misexpression of AtMYB36 in the root epidermis led to lignin and suberin deposition in the epidermis[11]. The latest research found that SHORT-ROOT (SHR) activates a hierarchical network for lignin and suberin formation. As a key hub regulator, AtMYB36 is located in the second layer and regulates multiple downstream AtMYB genes in the network. Interestingly, AtMYB6, AtMYB122 and AtMYB36 form a feed-forward loop. The AtMYB36 mutant destroyed the AtMYB36-AtMYB122-AtMYB6 feed-forward loop, weakened the inhibition of the downstream suberin genes, and thus led to an increase in endodermal suberization[14]. A recent study also showed that AtMYB36 plays a critical role in limiting cell division at the late stages of lateral root primordium development by regulating PER genes that regulate reactive oxygen species (ROS) balance and cell proliferation potential[13]. In rice, knockout of all three OsMYB36s led to the complete absence of the Casparian strip in the endodermal cell layer and inhibited plant growth[7].
Interestingly, CsMYB36 was reported to be involved in the formation of yellow–green peel in cucumber in addition to regulating root development[15]. MYB36 has also been found to play an important role in other secondary metabolic pathways. For example, scientists have found that ScMYB36 and other key transcription factors may be involved in the regulation of tanshinone synthesis by using comparative genomics, RNA sequencing and weighted gene coexpression network analysis (WGCNA) in Salvia castanea[16]. Consistent with recent results, SmMYB36 was reported to promote the accumulation of tanshinone by activating tanshinone biosynthesis enzyme genes (SmERF6 and SmCPS1) directly in Salvia miltiorrhiza. Similarly, SmMYB36 inhibited the accumulation of phenolic acid by directly repressing the expression of SmERF115 and SmRAS, genes involved in the synthesis of phenolic acid[17]. Plant MYB36 genes also play critical roles in the response to biotic and abiotic stresses. For example, in cotton, GhMYB36 positively regulates drought stress tolerance and Verticillium wilt resistance by enhancing PR1 expression[18]. In addition, the latest research found that AtMYB36 is likely to participate in cold adaptation and plant growth through metabonomics, transcriptomics, phenotype omics and GWAS[19]. All the studies indicated that the MYB36 family genes have multiple functions in the life cycle of plants.
Cucumber (Cucumis sativus L.) is one of the most important vegetable crops worldwide. With high-quality genome data[20, 21], a number of gene families have been identified, such as CsTCP[22], CsWOX[23], and lectin receptor-like kinases (LecRLKs)[24]. However, the identification of the R2R3-type CsMYB36 gene family has not been conducted, which greatly limits the study of the biological function of CsMYB36s in cucumber. Only one study showed the function of CsMYB36 in cucumber[15]. Root development is very important to the growth and development of plants. However, the detailed mechanism of cucumber primary root development remains obscure. Much research has focused on the growth and development of shoot branching, leaves, flowers and fruits in cucumber[25–33]. Thus, this study aimed to expand our understanding of the structure and function of the CsMYB36 gene family in cucumber. In our research, we identified 15 CsMYB36s in the cucumber genome. The comprehensive expression pattern of CsMYB36 in different tissues and in the root differentiation zone showed significant differential expression of CsMYB36s and proposed putative CsMYB36 gene members related to root development. Our results also suggested that CsMYB36s may play a critical role in the response to high temperature and waterlogging stress.
-
We selected plants with different evolutionary histories to analyse the evolution of CsMYB36. These species included dicotyledons, namely, Amborella trichopoda (AMTR), Nymphaea colorata (NC), Glycine max (Glyma), Arabidopsis thaliana (AT), Solanum lycopersicum (Solyc), and Cucumis sativus (Cs); a monocotyledons, namely, Zea mays (Zm) and Oryza sativa (Os); a gymnosperm, Ginkgo biloba (Gb); a moss, Physcomitrella patens (Pp); and a fern, Selaginella moellendorffii (Smo). The amino acid sequences of plants were obtained from Phytozome v12. The amino acid sequences of AtMYB36 were used as a query to search for MYB36 proteins in all the plant species’ protein files by using TBtools software, and multiple sequence alignment was performed by MEGA 11. The phylogenetic trees were constructed by the Phylogeny program of MEGA 11 using the neighbour-joining (NJ) model and the Shimodaira–Hasegawa test.
The exon/intron structures of the MYB36 genes identified in Cucumis sativus and Arabidopsis thaliana were generated using TBtools. The conserved motifs in the MYB36 proteins identified in Cucumis sativus and Arabidopsis thaliana were analysed with MEME software[8]. The conserved motifs of MYB36 were annotated with the Pfam database. Multiple alignments of the MYB36 protein sequences in cucumber and Arabidopsis were performed by Muscle in MEGA 11 with default parameters. The phylogenetic trees were constructed by the Phylogeny program of MEGA 11 using the NJ model and Shimodaira–Hasegawa test.
Analysis of synteny with other plants
-
TBtools software was used to analyze and visualize the syntenic relationships of the MYB36 genes in cucumber, Arabidopsis, and rice.
Analysis of cis-acting elements of CsMYB36 gene promoters
-
The cis-elements in the promoter sequence (2,000 bp DNA sequence upstream of the start codon) of all CsMYB36s were analyzed using the PlantCARE database[11].
Expression profile analyses of CsMYB36 genes based on RNA-seq
-
For the expression pattern analysis of CsMYB36 genes in different tissues of cucumber, we obtained the RNA-seq data for cucumber gene expression in different tissues from the cucumber genomic database (data number: PRJNA80169). To explore the expression patterns of the CsMYB36 genes in the different zones of the root, the RNA-seq data available in the cucumber genomic database were retrieved (data number: PRJNA271595). In order to understand the expression of CsMYB36 genes in response to various abiotic stresses, public RNA-seq data were retrieved from the NCBI SRA database. Such as, GA treatment (data number: PRJNA376073) and temperature and photoperiod treatment (data number: PRJNA380322). TBtools software was used to draw the the expression heatmap of CsMYB36s in cucumber.
Expression pattern of cucumber CsMYB36 genes in response to hormone stresses
-
According to cis-acting regulatory elements prediction, we choose Auxin (IAA), Abscissic acid (ABA) and Methyljasmonate (MeJA) to treat the whole plant of cucumber. Cucumber seeds were soaked for 30 min at 55 °C and the seed shell removed. Seeds were sterilized with 75% alcohol for 30 s, and seedlings were grown on 1/2 Murashige & Skoog medium (MS) at 25 °C under 14-h day/10-h night for 4 d. For hormone treatment, uniform-sized seedlings were divided into four groups, transplanting in IAA 1 uM + 1/2 MS, ABA 20 uM + 1/2 MS, MeJA 0.5 uM + 1/2 MS and 1/2 MS. Seedlings were grown in a growth room at 25 °C under 14-h day/10-h night for 1 d, and then we collected the roots (three repeats for each treatment) for qRT-PCR analysis. The primers are shown in Supplemental Table S1.
Expression pattern of cucumber CsMYB36 genes in response to waterlogging
-
After 7 d of cultivation, plants were divided into two groups, control and stress groups. Before stress treatment, the percent volumetric water content (VWC) was measured and plants were watered to obtain a soil moisture level up to 30%. The root zone of cucumber plants from the stress group were waterlogged for 7 d in a deep tray containing water.
-
AtMYB36 regulates the whole root development by inhibiting cell differentiation. Our team found that CsMYB36 is involved in the formation of yellow green peel in cucumber. Thus, we speculate that there are multiple CsMYB36s in cucumber, and their functions are differentiated during evolution. To understand the evolution and conservation of CsMYB36, we analyzed the amino acid sequence of CsMYB36 proteins in cucumber genomes and constructed a phylogenetic tree using the amino acid sequences (Fig. 1). The phylogenetic trees indicated that the high conservation of most MYB36 genes during the evolution among different plant species. We identified 15 members of the CsMYB36 gene family in cucumber according to the phylogenetic trees (Supplemental Table S2 & Supplemental Table S3). Supplemental Table S2 shows the detailed data about these CsMYB36s, including the gene ID, length of protein, isoelectric point, molecular weight (MW), e-value, etc. The length of the identified CsMYB36 proteins varied from 204 amino acids (CsaV3_2G025790.1 and CsaV3_5G038180.1) to 387 amino acids (CsaV3_6G049630.1). The isoelectric point varied from 5.74 (CsaV3_1G011720.1) to 9.17 (CsaV3_1G013560.1), and the MW varied from 23037.2 Da to 44518.01 Da.
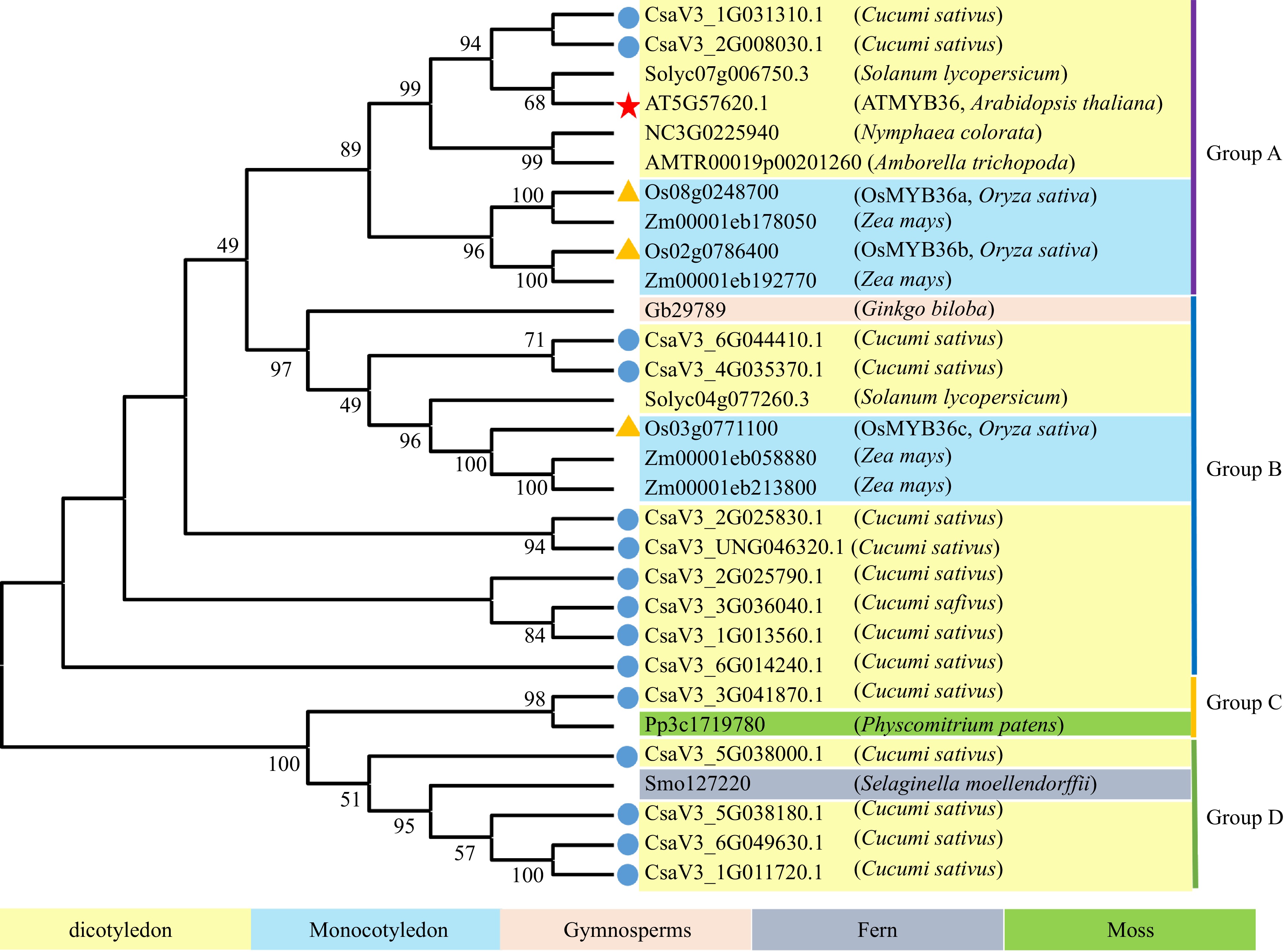
Figure 1.
Phylogenetic analysis of CsMYB36 homologues in Cucumis sativus and other plants. Blue dots highlight CsMYB36 homologues in Cucumis sativus. Yellow triangles highlight MYB36 homologues in Oryza sativa. The scale bar is the number of amino acid substitutions per site. See Supplemental Table S2 for the alignments of MYB36 homologues in different species.
We divided the evolutionary tree into four subgroups: A, B, C and D based on the structural features of their protein sequences. Based on phylogenetic analysis, we found that ten CsMYB36 genes of cucumber in group A and B have higher homology with that of tomato, Arabidopsis, maize and rice (Fig. 1). It has been revealed that MYB36 plays an important role in the deposition of Casparian strip and the transport of apoplast in Arabidopsis and rice roots. These results indicated that the ten CsMYB36 proteins may function in root development. However, one MYB36 proteins (CsaV3_3G041870.1) of group C is closely homologous to the Pp3c1719780 protein which belongs to the moss (Physcomitrium patens). The moss plants lack roots. Therefore, it is suggested that CsaV3_3G041870.1 may play the role in the growth and development of other tissues and organs. Four MYB36 proteins of group D are closely homologous to the fern (Selaginella moellendorffii).
The classification, motif composition, and gene structure of the CsMYB36 gene family
-
To understand the conserved structure of MYB36 proteins, we drew the motif distribution by using the MEME and TBtools software. To classify the CsMYB36 genes, two neighbor-joining phylogenetic trees were constructed by using the MYB36 protein sequences or MYB36 protein sequences from Arabidopsis. Based on the support of bootstrap value > 50%, MYB36 proteins from cucumber could be divided into two groups (designated A and B) (Fig. 2).
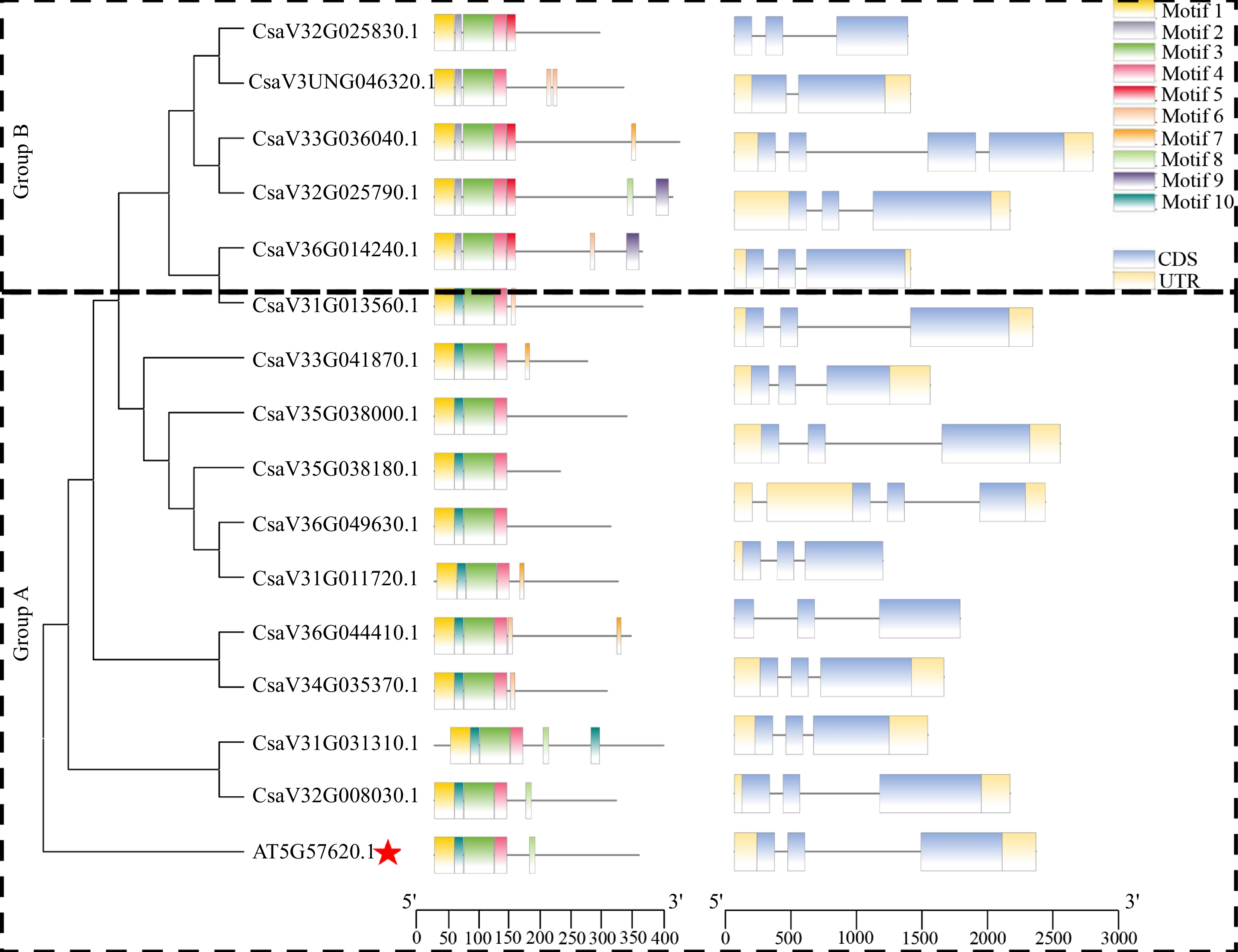
Figure 2.
Phylogenetic tree, conserved motifs and gene structure of CsMYB36 family genes in Arabidopsis thaliana and Cucumi sativus. The red star highlights the MYB36 homolog in Arabidopsis thaliana.
According to coding sequences of CsMYB36s in cucumber, we found that the gDNA lengths of CsMYB36 varied from 1132 (CsaV3_6G014240.1) to 2736 bp (CsaV3_5G038180.1), and the CDS lengths ranged from 612 (CsaV3_2G025790.1) to 1161 bp (CsaV3_6G049630.1) (Supplemental Table S2). The number of introns of these CsMYB36 genes ranged from one to three. The number of exons in CsMYB36 was quite similar (2 – 4 per gene), which is similar to the structure of the CsMYB36 family in Arabidopsis genomes, possibly indicative of functional redundancy (Fig. 2). Ten conserved motifs named Motif 1 to Motif 10 were identified in CsMYB36s. The logo and name of the query motifs are displayed in Supplemental Table S4. The different colored boxes in the legend referred to distinct motifs. Motif 1, 3 and 4 (the yellow, green, and pink motifs) are present in all the CsMYB36 proteins and represented the conserved MYB36 domain. However, motifs 10 (dark green motif) exist in the group A. Proteins containing only motif 5 included CsaV3_2G025830.1, CsaV3_3G036040.1, CsaV3_2G025790.1 and CsaV3_6G014240.1 were from the group B. Similar to motif 5, motif 2 only exists in group B. The amino acid sequence of all 15 homologs had the R2R3-MYB conserved domains of typical MYB proteins (Supplemental Fig. S1). These results revealed that most CsMYB36 proteins from the same group had similar motif compositions, and indicated that the proteins from the same group may own a similar function.
Collinearity analysis of the CsMYB36 family genes
-
We constructed the comparison system diagram between cucumber and other two plants to explore the interspecifc collinearity of the CsMYB36 family in cucumber and other species (Fig. 3). Analysis of collinearity showed that there were 19 collinearity gene pairs between cucumber with Arabidopsis genome. There was no collinearity gene pair on Arabidopsis Chr1 with any putative CsMYB36 genes in the cucumber genome. Five cucumber CsMYB36s and four rice OsMYB36s were orthologous genes with five syntenic relationships. Two cucumber CsMYB36 genes CsaV3_1G013560.1 and CsaV3_3G036040.1 did not form the syntenic gene pair with neither A. thaliana nor rice, which suggested that these two genes are relatively conservative during evolution (Supplemental Table S5).
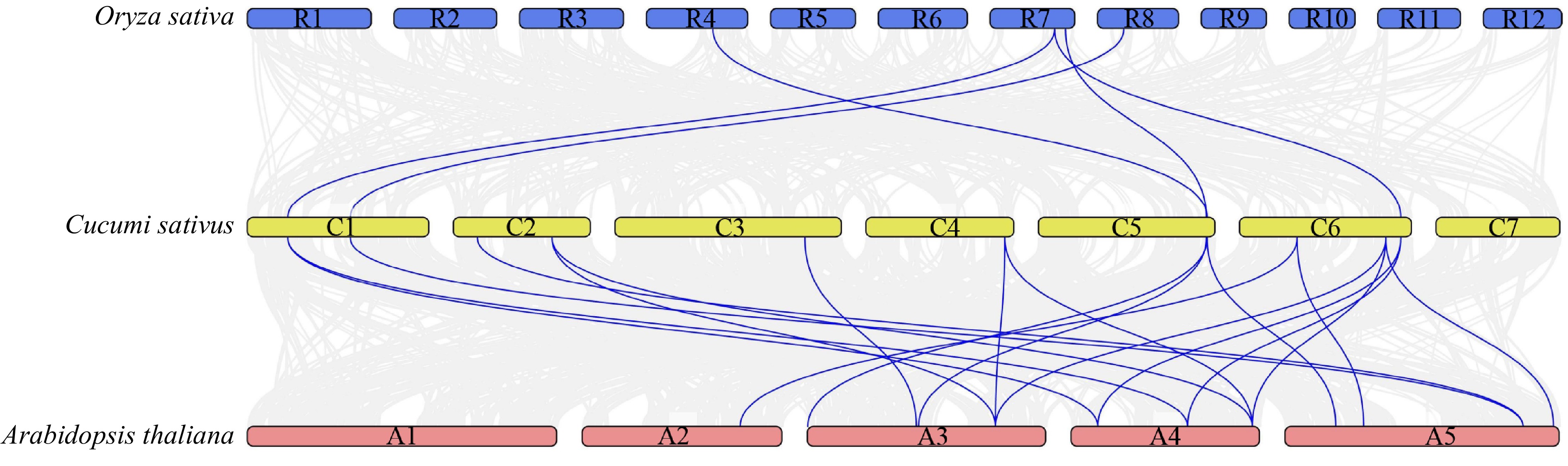
Figure 3.
Syntenic analysis of MYB36 family genes in Cucumis sativus, Arabidopsis thaliana and Oryza sativa. The blue line highlights the genes with syntenic relationships. See Supplemental Table S5 for these genes ID.
Analysis of the cis-acting element in the promoter of CsMYB36s
-
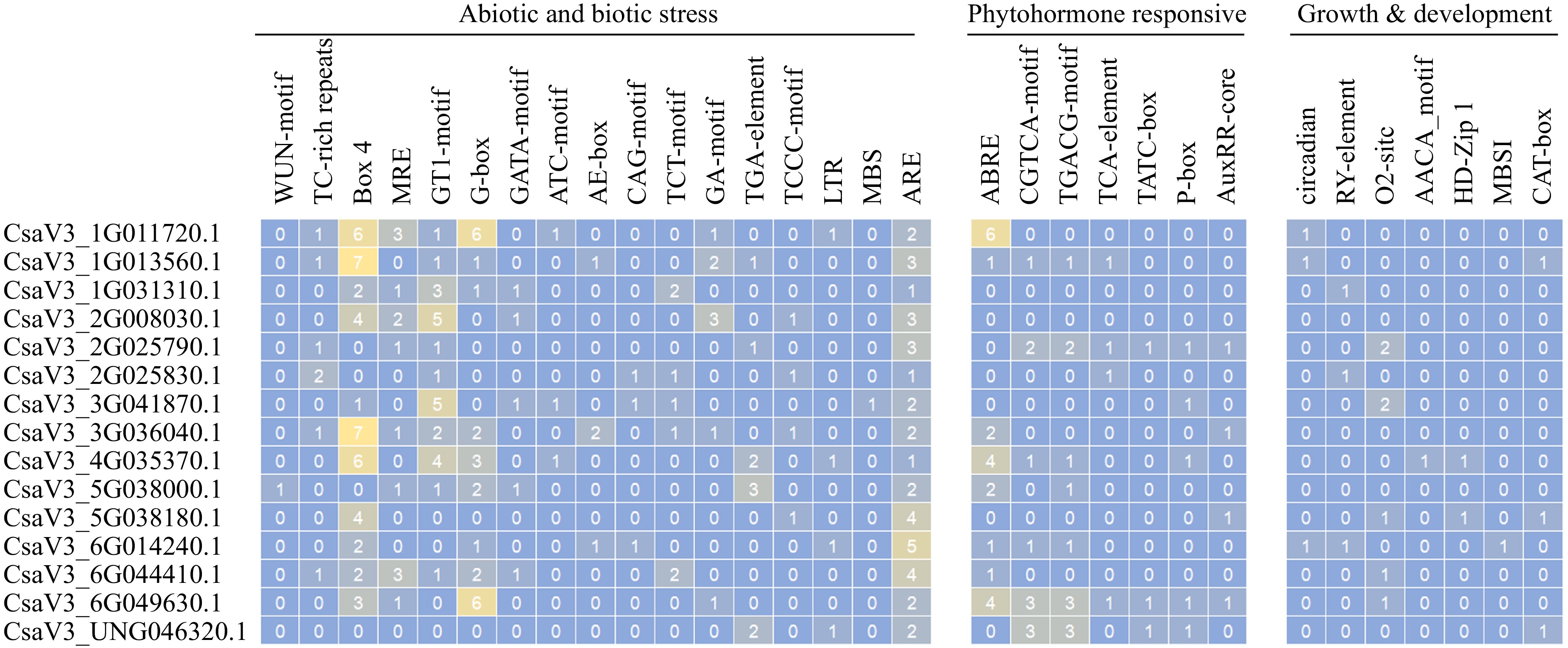
To investigate the potential biological functions of CsMYB36, the promoter sequences of 15 CsMYB36s were analyzed by the PlantCARE database[34]. As a result, a total of 17 main types of cis-acting elements were identified and are shown in Fig. 4[34]. Cis-regulatory elements related to abiotic and biotic stress, phytohormone regulation, and plant growth and development were determined in the promoters of the 15 CsMYB36s (Fig. 4 & Supplemental Tables S6, S7). Previously reported showed that bHLHs and bZIPs domain protein bind to the G-box (CACGTG) cis-acting element and regulate the response of plants to stress[35]. We found that most promoters of CsMYB36 gene contain G-box elements. Similarly, the GT-1 motif was found in most CsMYB36s promoters (except CsaV3_5G038180.1, CsaV3_6G014240.1, CsaV3_6G049630.1 and CsaV3_UNG046320.1). It has been reported that the GT-1 motif was linked to salt and osmotic stresses in Arabidopsis thaliana[36]. One WUN-motif (involved in wound response) elements were present in CsaV3_5G038000.1, suggesting that it has important functions in response to wound stress. Interestingly, all CsMYB36s are found to have anaerobic-induction (ARE motif) elements (Supplemental Tables S6, S7).
Phytohormonal cis-acting elements involved in the regulation of MeJA-responsive element (CGTCA motif and TGACG motif)[37] were found to be enriched in the promoters of CsaV3_1G013560.1, CsaV3_4G035370.1, CsaV3_6G014240.1 and CsaV3_6G049630.1. SA-responsive element -TCA element[38], were found to be enriched in the promoters of CsaV3_1G013560.1, CsaV3_2G025790.1, CsaV3_6G049630.1 and CsaV3_2G025830.1 genes. As expected, the ABA-responsive element-ABRE[39] was highly enriched in a large number of CsMYB36 genes (CsaV3_1G011720.1, CsaV3_1G013560.1, CsaV3_3G036040.1, CsaV3_4G035370.1, CsaV3_5G038000.1, CsaV3_6G014240.1, CsaV3_6G044410.1, and CsaV3_6G049630.1) (Fig. 4). This indicates the probable cross-wired mesh of ABA signaling in plant development processes through the involvement of CsMYB36s. Other phytohormone-related cis-regulatory elements were identified. For instance, auxin response elements AuxRP elements[40] and gibberellin response elements TATC-box and P-box[41] in the promotors of some CsMYB36s (Fig. 4 & Supplemental Tables S6, S7).
For growth and development-related cis-regulatory elements, we found the circadian control-related cis-regulatory elements circadian and the seed specific regulation element (RY-element) existed in CsaV3_1G011720.1, CsaV3_1G013560.1 and CsaV3_6G014240.1, and in CsaV3_1G031310.1, CsaV3_2G025830.1 and CsaV3_6G014240.1, respectively. The zein metabolism regulation and endosperm expression-related elements AACA-motif were enriched in the promoter of CsaV3_2G025790.1, CsaV3_3G041870.1, CsaV3_5G038180.1, CsaV3_6G044410.1 and CsaV3_6G049630.1 and in CsaV3_4G035370.1, respectively. Additionally, HD-Zip1 (differentiation of the pallisade mesophyll cells-related) was present in CsaV3_4G035370.1 and CsaV3_5G038180.1, and O2 site motifs (zein metabolism regulation-related) was present in CsaV3_2G025790.1, CsaV3_3G041870.1, CsaV3_5G038180.1 and CsaV3_6G049630.1, respectively. The different members of cis-acting elements existed in the promoter position of different CsMYB36s, revealing that CsMYB36s are important in the process of plant growth and development in cucumber.
Expression analysis of CsMYB36 genes in different plant tissues
-
We examined the expression pattern of CsMYB36 family genes in different tissues of cucumber according to the published RNA-seq data (Fig. 5a)[42]. Four CsMYB36 genes, CsaV3_1G031310.1, CsaV3_6G044410.1, CsaV3_4G035370.1 and CsaV3_2G008030.1 were only highly expressed in root indicating that the four genes function in the root development. Notably, the gene of CsaV3_2G025830.1 highly expressed in the root and ovary tissues. These results suggest that CsaV3_2G025830.1 function in the fruit and root development. Indeed, consistent with the transcriptional results, a previous study showed that CsaV3_2G025830.1 was involved in the formation of yellow green peel in cucumber[15]. CsaV3_3G036040.1 was most expressed in root and tendril. CsaV3_2G025790.1 was special highly expressed in the ovary tissue, indicating that CsaV3_2G025790.1 plays an critical role in the fruit set. The transcript levels of CsaV3_5G038180.1 and CsaV3_5G038000.1 were consistently high in every organ except root. In contrast, CsaV3_6G014240.1, CsaV3_3G041870.1 and CsaV3_1G011720.1 were not detected in most organs, indicating that these three CsMYB36s maybe the pseudogenes. CsaV3_6G049630.1 was only expressed in the root and the transcript level was low. CsaV3_UNG046320.1 had the strongest expression in female flower, male flower and root. Similarly, CsaV3_1G013560.1 had the strongest expression in root, ovary, and leaf.
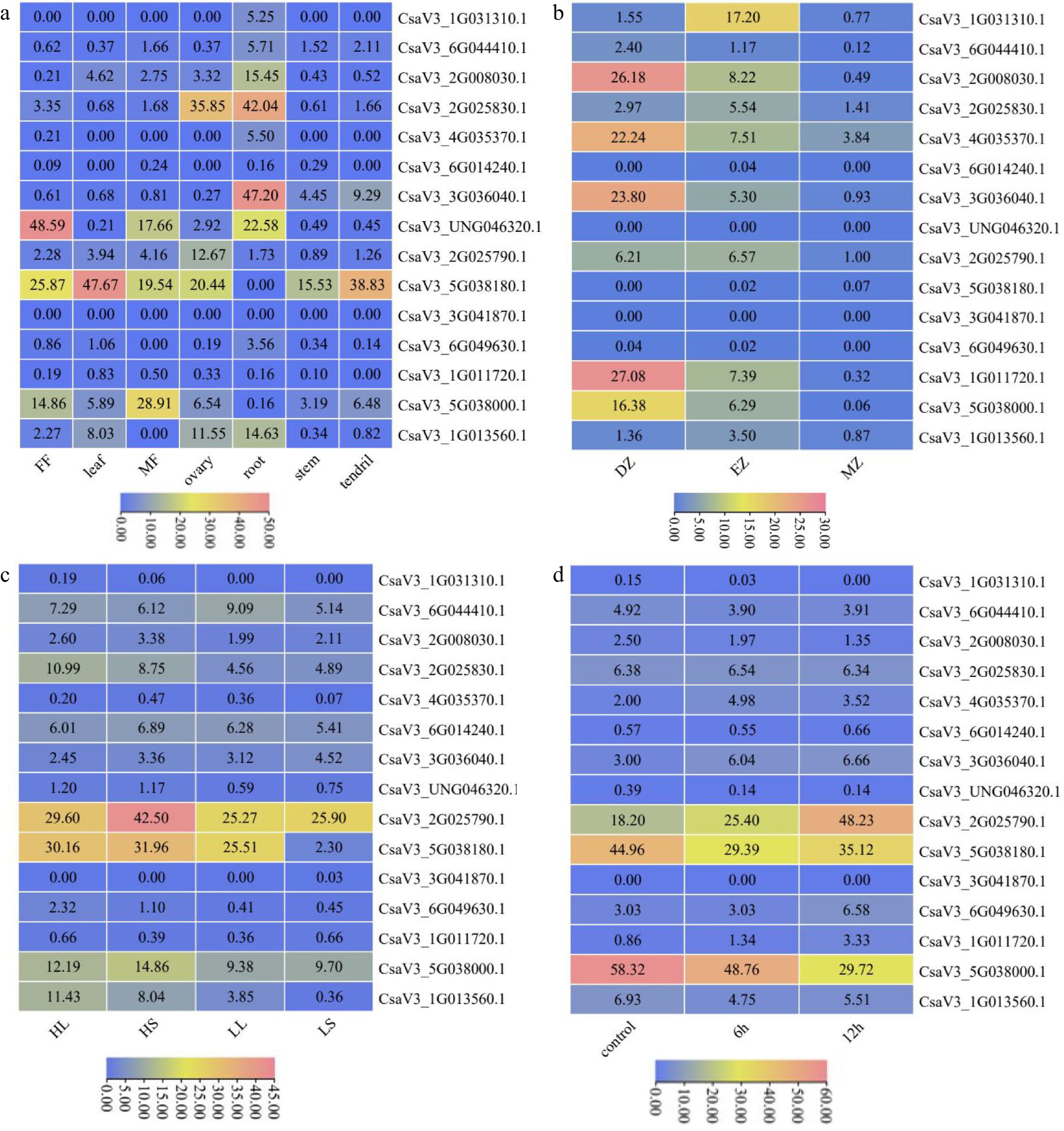
Figure 5.
Expression patterns of CsMYB36 genes. (a) Expression heatmap of CsMYB36s in different tissues of cucumber. (b) Expression heatmap of CsMYB36s under different temperature and photoperiod treatments. HighTemp, LongPhoto (HL); HighTemp, ShortPhoto (HS); LowTemp, LongPhoto (LL); LowTemp, ShortPhoto (LS). (c) Expression heatmap of CsMYB36s in different root zones. Root differentiation zone (DZ); root elongation zone (EZ); root meristematic zone (MZ). (d) Expression heatmap of CsMYB36s under different GA treatments. Cells were treated with GA at 6 h and 12 h.
Expression analysis of cucumber CsMYB36 genes in roots
-
Since CsMYB36 was reported to play a key role in root development, we examined the transcript pattern of CsMYB36s in the different zones of the whole root. Based on the published cucumber RNA-seq data in the root differential zones[42], similarly, we drew the expression heatmap of the cucumber CsMYB36 gene family in the root (Fig. 5b). Five CsMYB36 genes, CsaV3_2G008030.1, CsaV3_3G036040.1, CsaV3_4G035370.1, CsaV3_1G011720.1 and CsaV3_5G038000.1 had the strongest expression in the root differentiation zone and had the moderate expression in the root elongation zone. It indicates that these CsMYB36 genes may function in root differentiation. In contrast, CsaV3_6G044410.1, CsaV3_6G014240.1, CsaV3_UNG046320.1, CsaV3_5G038180.1, CsaV3_3G041870.1, CsaV3_6G049630.1 and CsaV3_1G013560.1 were almost not detected in the whole root, suggesting that these genes probably have no function in the root. CsaV3_1G031310.1 were only highly in the root elongation zone. CsaV3_2G025830.1 was expressed in the whole root, however, the expression of CsaV3_2G025830.1 in the root was low. Combined with the results of tissue-specifc expression analysis, CsaV3_2G008030.1 and CsaV3_3G036040.1 may function in the formation of root CS.
Expression analysis of CsMYB36 genes under temperature and photoperiod treatment
-
We checked the expression pattern of the cucumber CsMYB36 gene family under temperature and photoperiod treatment based on the published cucumber RNA-seq data[43] (Fig. 5c). We found only one gene, CsaV3_1G013560.1 was significantly up-regulated under high temperature stress in both long and short days. CsaV3_ 2G025830.1 was up-regulated by high temperature under long days. The expression levels of the CsaV3_5G038180.1 at high temperature and short day were high than that at low temperature and short day, indicating that the CsaV3_5G038180.1 gene responded to high temperature stress under short day. In addition, the transcript levels of CsaV3_5G038180.1 and CsaV3_1G013560.1 under low temperature and long day treatment were higher than those under low temperature and short day treatment, which revealed that these two CsMYB36 genes were involved in photoperiod response (Fig. 5c).
Expression pattern of CsMYB36 genes under GA treatment
-
We plotted the expression heatmap of the cucumber CsMYB36 gene family under GA treatment according to the published RNA-seq data[44] (Fig. 5d). The expression levels of CsaV3_1G011720.1 and CsaV3_2G025790.1 were up-regulated at 6 h and 12 h after GA treatment. The expression level of CsaV3_4G035370.1 was up-regulated under GA treatment at 6 h when compared with the control. Similarly, the expression level of CsaV3_6G049630.1 was up-regulated under GA treatment at 12 h. These results indicated that only these cucumber CsMYB36 family genes were closely related to GA signal transduction.
Expression analysis of CsMYB36 genes under phytohormone treatment
-
Auxin and ABA play a critical role in the development of root. Thus, we conducted Auxin and ABA treatment on cucumber roots. Moreover, it was found that most of the CsMYB36 promoters exist JA binding elements based on the analysis of promoters cis-elements of the CsMYB36 and we speculated that whether CsMYB36 played a role in the JA mediated defense process. Therefore, in order to further verify the response of CsMYB36 to JA, we also conducted JA treatment on cucumber. We selected the top five genes homologous to Arabidopsis CsMYB36 according to the e-value (Supplemental Table S2), and checked their expression levels of CsMYB36s under IAA, ABA, and JA treatment. As shown in Fig. 6, the transcript levels of CsaV3_2G025830.1 and CsaV3_4G035370.1 increased significantly under IAA treatments. The expression levels of CsaV3_6G044410.1 and CsaV3_2G008030.1 increased significantly under ABA treatments. However, the expression level of CsaV3_2G025830.1 decreased significantly under ABA treatments. Interestingly, there was no significant change in the transcript of all five genes under JA treatment indicating that the five genes don’t respond to JA (Fig. 6).
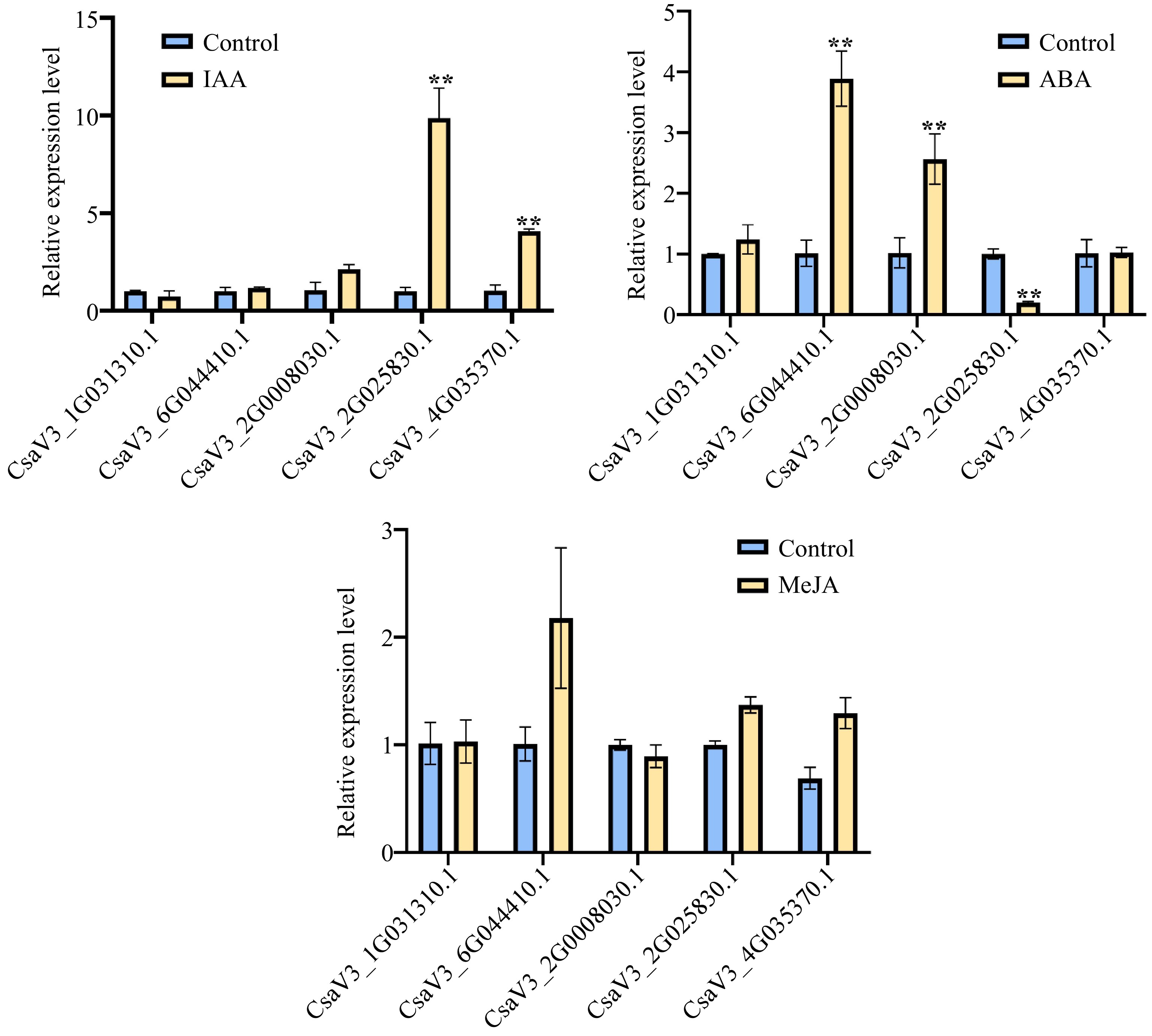
Figure 6.
Relative expression levels for CsMYB36s. (a) Relative expression levels of five cucumber CsMYB36s after 24 h 1 uM 3 Indoleacetic acid (IAA) treatment in cucumber seedlings by qRT -PCR analysis. (b) Relative expression levels of five cucumber CsMYB36s after 24 h 20 uM Abscisic acid (ABA) treatment in cucumber seedlings by qRT -PCR analysis. (c) Relative expression levels of five cucumber CsMYB36s gene after 24 h 0.5 uM Methyljasmonate (MeJA) treatment in cucumber seedlings by qRT-PCR analysis. Data are displayed using the CsActin as an internal control with three biological and three technical replicates. Values are the mean ± SD. Significant differences were observed between treatments (**P < 0.01; *P < 0.05, Student's t test).
Expression analysis of CsMYB36 genes under waterlogging stress.
-
It was found that all the CsMYB36s promoters contain the cis-acting regulatory element essential for anaerobic induction. Therefore, in order to further verify the response of CsMYB36 to waterlogging, we also conducted waterlogging treatment on cucumber seedling. As shown in Fig. 7, as we expected, 13 genes responding to waterlogging stress. Among them, eight genes were up-regulated and five genes were down-regulated by waterlogging stress (Fig. 7).
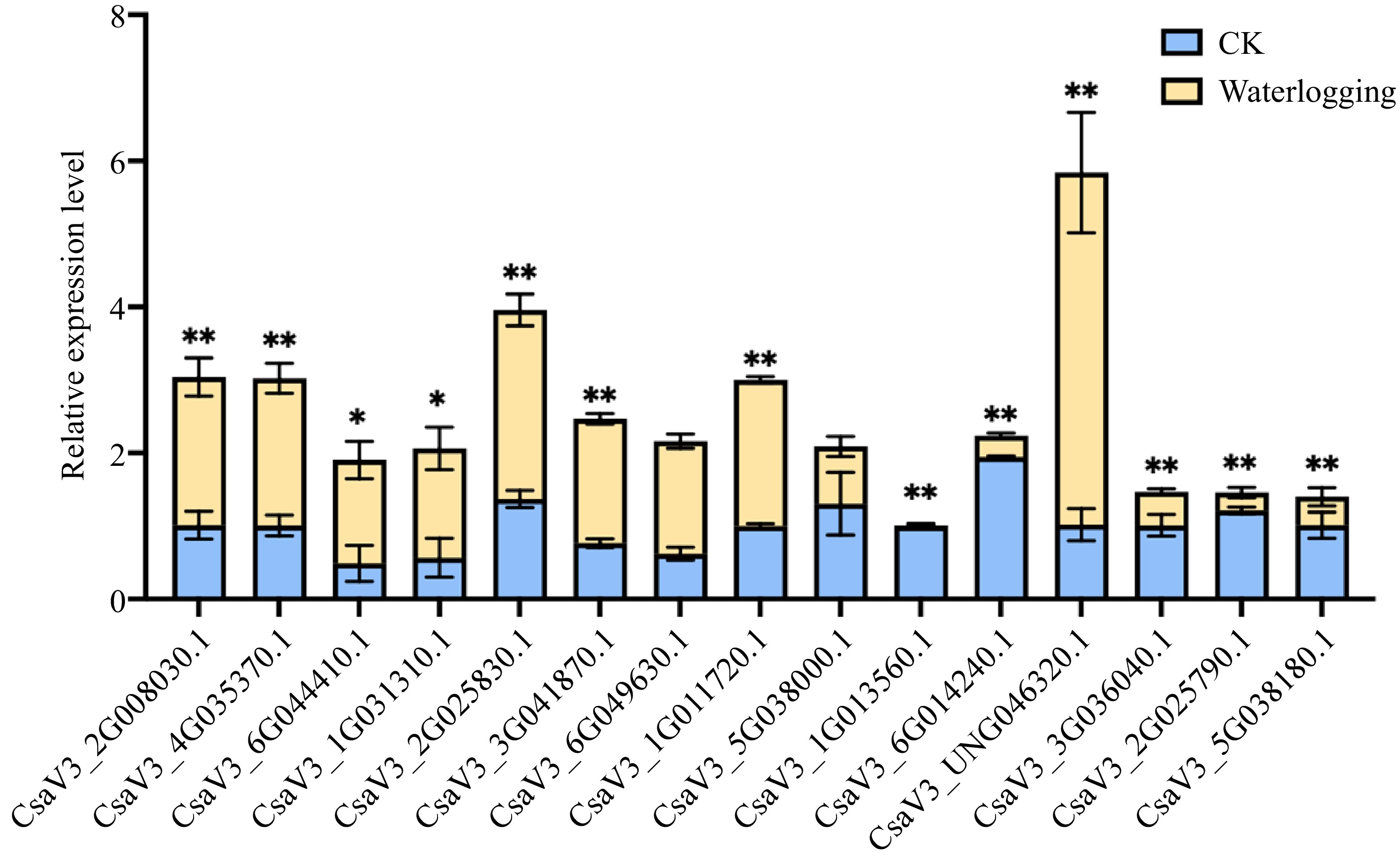
Figure 7.
Relative expression levels for CsMYB36s under waterlogging stress. Relative expression levels of CsMYB36s after 7 d waterlogging treatment in cucumber seedlings by qRT-PCR analysis. Data are displayed using CsActin as an internal control with three biological and three technical replicates. Values are mean ± SD. Significant differences were observed between treatments (**P < 0.01; *P < 0.05, Student's t test).
-
Plants mine the surrounding soil for water and dissolved minerals to sustain growth and complete their life cycle. Cell wall lignification is essential for forming functional endodermis in the mature zone of plant roots. The lignified apoplastic structure in the endodermis is called the Casparian strip (CS), which is critical for the selective absorption of water and nutrients in plant roots[19]. Cucumber (Cucumis sativus L.) is an important vegetable crop with great economic value. Many important discoveries have been made regarding different developmental processes in cucumber[25−33]. However, there are few reports on the genes related to cucumber root development. In recent years, multiple key components of the molecular machinery promoting the formation of the CS have been identified, among which SHORT-ROOT and its downstream R2R3 type family gene MYB36 are the main regulatory factors[8−11]. A previous study showed that loss-of-function myb36 mutants have delayed and defective barrier formation in the Arabidopsis root endodermis[8−9]. In rice, knockout of all three OsMYB36 genes led to the complete absence of the CS in the endodermal cell layer and inhibited plant growth[7]. These results showed that the function of MYB36 in controlling the formation of the CS in plants is probably conservative. Interestingly, the transcription factor MYB36 is essential for yellow‒green peel formation in cucumber[15]. These results suggest that MYB36 not only plays a critical role in the formation of CS in the root but also functions in the development of fruit. Thus, we speculate that there are multiple CsMYB36s in cucumber, and their functions may be differentiated during long-term evolution. To mine MYB36s with functions in the root, we identified the CsMYB36 family genes in cucumber and analysed the expression patterns of CsMYB36s in different tissues and in response to different hormone treatments. Fifteen CsMYB36 family genes were identified in cucumber. Phylogenetic tree analysis of 15 CsMYB36s derived from dicotyledons, monocotyledons, a gymnosperm, a moss and a fern was divided into four subgroups: A, B, C, and D. Ten of the cucumber CsMYB36 homologues are closely related to AtMYB36, SlMYB36 and OsMYB36[8−10,12]. Genes with similar functions are clustered together in the phylogenetic tree. To date, MYB36 has been reported to be the key transcription factor regulating the formation of the CS in Arabidopsis, rice and tomato roots, which led us to hypothesize a similar function of the ten CsMYB36 genes in the maturation of the endodermis. Indeed, except for CsaV3_6G014240.1, the other nine CsMYB36 genes were highly expressed in roots. Among these nine CsMYB36 genes, CsaV3_2G025830.1 has been identified in cucumber. The peel of cucumber fruit is yellow–green in the CsaV3_2G025830.1 mutant plant. Consistent with the observed phenotypes, CsaV3_2G025830.1 is also highly expressed in the cucumber ovary[15]. This implies that subfunctionalisation of CsMYB36 homologues may have occurred during cucumber evolution. In addition, CsaV3_2G008030.1 and CsaV3_3G036040.1 were highly expressed in the root differentiation zone (Fig. 5b), which is consistent with the position of the CS in the root. Considering that CsaV3_2G008030.1 and CsaV3_3G036040.1 are only highly expressed in the root, we speculate that CsaV3_2G008030.1 and CsaV3_3G036040.1 may play a key role in the formation of the CS in cucumber root. Mosses have no roots, and we found that only one CsMYB36 (CsaV3_3G041870.1) paralogue was closely related to moss. However, this gene is not expressed in any cucumber tissue, indicating that the gene probably has no function in cucumber. A number of studies have reported that R2R3-type MYBs function in flower development[45−47]. We found that the transcript levels of CsaV3_5G038000.1 from subgroup D were higher in male flowers and female flowers than in other tissues, which indicates that CsaV3_5G038000.1 may play important roles in regulating flower development in cucumber. Interestingly, we found that CsaV3_5G038180.1 was not expressed in the root but was highly expressed in other tissues, implying that CsMYB36 may participate in a wide range of developmental processes in cucumber.
Through expression pattern analyses of cucumber CsMYB36 family genes in response to temperature and photoperiod stresses, only one CsMYB36, CsaV3_1G013560.1, was identified to respond to high temperature stress. This result was not found in the other plant MYB36 genes. This suggests that CsMYB36 also plays an important role in the response to high-temperature stress, which provides a new clue for the genetic improvement of high-temperature tolerance in cucumber. In addition, four cucumber CsMYB36s were upregulated after GA treatment. Among the four CsMYB36s, we found that the promoter sequences of three CsMYB36s presented GA response elements. Thus, it is believed that CsMYB36 genes are related to the response to GA stress. ABA does not affect the expression levels of AtMYB36 at either the transcriptional or translational levels[12]. However, we found that CsMYB36s respond to ABA treatment. In Arabidopsis thaliana, AtMYB74 and AtMYB68 homologues of AtMYB36 all respond to ABA, and their overexpression increases lignification and suberization of the root endodermis[16,48]. It is suggested that the three CsMYB36s expressed in roots may mediate ABA signalling-induced root lignification. Moreover, we found that nearly all the promoter sequences of CsMYB36 contain anaerobic induction elements. The high number of anaerobic induction element-specific inducibility would lead to the high expression of CsMYB36 family genes in anaerobic environments, such as waterlogging. An oxygen-deficient state will occur when plants are in a flooded state for a long time[49]. Research has shown that roots establish a lateral diffusion barrier to minimize radial oxygen loss from flooded roots to the soil. For example, Zea nicaraguensis (teosinte) is a wild relative of Zea mays (maize), but unlike maize, it is tolerant to waterlogging. Compared with maize roots, the deposition of lignin and suberin in the outer cell layers of teosinte roots is enhanced under flooded conditions, which increases the gas impermeability of the root surface, thus greatly reducing the radial loss of oxygen in teosinte[50]. Similarly, lignification and suberin deposition in the root exodermal cell layer of rice provide a barrier for radial oxygen loss[51]. Consistent with these observations, expression of CsMYB36s is highly significantly upregulated after waterlogging[52]. Therefore, we speculate that plants adapt to hypoxia by regulating the expression levels of CsMYB36s, thereby affecting formation under flooded conditions.
In conclusion, we identified 15 CsMYB36s in the cucumber genome. The comprehensive expression pattern of CsMYB36s in different tissues and in the root differentiation zone indicated significant differential expression of CsMYB36s and putative root development-associated gene members. Our study lays a foundation for further screening the MYB36 genes necessary for the development of the CS by reverse genetics and discovering possible new functions of CsMYB36s in other development or stress processes.
This work was funded by the National Key Research and Development Program (2022YFD1200502), National Natural Science Foundation of China (32211540386). Hunan Provincial Science and Technology Innovation Plan Program (2022RC1143), Hunan Provincial Natural Science Foundation of China (2022JJ40164) and Scientific Research Fund of Hunan Provincial Education Department (19B271).
-
The authors declare that they have no conflict of interest.
-
# These authors contributed equally: Chunhua Wang, Xi Shen
- Supplemental Fig. S1 Sequences of all CsMYB36 members in Cucumis sativus. Sequence alignment of cucumber CsMYB36s, Arabidopsis MYB36 using DNAMAN.
- Supplemental Table S1 The primers used for qRT-PCR of CsMYB36s in this study.
- Supplemental Table S2 Detail information description of the CsMYB36 genes in Cucumber (Cucumis sativus L.).
- Supplemental Table S3 Protein sequence of CsMYB36s.
- Supplemental Table S4 The logo and name of motifs of CsMYB36s in this study.
- Supplemental Table S5 Syntenic relationships of MYB36 genes family in cucumber, Arabidopsis, and rice.
- Supplemental Table S6 The cis-elements of the CsMYB36s genome sequence (detail information).
- Supplemental Table S7 The cis-elements of the CsMYB36s genome sequence (data analysis).
- Copyright: © 2023 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Wang C, Shen X, Yang T, Yao H, Peng X, et al. 2023. Genome-wide characterization and identification of root development and stress-related CsMYB36 genes. Vegetable Research 3:19 doi: 10.48130/VR-2023-0019
Genome-wide characterization and identification of root development and stress-related CsMYB36 genes
- Received: 28 December 2022
- Accepted: 03 April 2023
- Published online: 14 June 2023
Abstract: Cucumber (Cucumis sativus L.) is an important vegetable crop worldwide. Over the last two decades, there have been many breakthroughs in the understanding of various developmental processes in cucumber, such as shoot branching, sex differentiation, and leaf and fruit size and shape. Roots play an important role in plant growth and development and nutrient absorption, affect crop yield, and participate in the interaction between plants and abiotic stress signals. However, the discovery of essential genes in cucumber roots is very limited. MYB36 is a critical transcription factor for whole development that negatively regulates cell proliferation and thus orchestrates Casparian strip (CS) formation in the root endodermis. To identify CsMYB36 genes with functions in the CS of cucumber and their responses to stress, we described in detail the characteristics of the CsMYB36 gene family in cucumber in this study. A total of 15 CsMYB36 genes were found in the cucumber genome. Through phylogenetic and tissue-specific expression analysis, CsaV3_2G008030.1 and CsaV3_3G036040.1 were selected as candidates regulating CS formation. In addition, all CsMYB36s were found to have anaerobic induction elements, indicating that MYB36 is likely to be highly responsive to anaerobic environments, such as waterlogging. We also identified one potential candidate CsMYB36 (CsaV3_1G013560.1) that participates in high-temperature stress responses. This work provides valuable information for understanding the characteristics of the CsMYB36 gene family in cucumber and provides new clues for researchers to study high temperature, waterlogging stress and the formation of CS.
-
Key words:
- Cucumber /
- CsMYB36 /
- Expression profles /
- Gene family