-
Eggplant (Solanum melongena. L) in the Solanaceae family is a globally and economically important vegetable crop rich in nutrition[1−4]. The color of eggplant fruits, primarily purple, white, or green, is an important quality and commercial trait[5−7]. Purple color varieties, containing high levels of anthocyanins in the fruit peel, are commonly found in the market[8,9]. As a branch of flavonoids in secondary metabolites, anthocyanin is a water-soluble natural pigment responsible for the blue, purple, and red color of many plant tissues[10−12].
The biosynthesis pathway of anthocyanin has been extensively studied in many plant species[13−16]. Both structural and regulatory genes have been found to participate in anthocyanin biosynthesis. Structural genes encode enzymes that directly catalyze anthocyanin biosynthesis, while regulatory genes mainly refer to those coding for transcription factors (TFs) that manipulate the expression of structural genes[17−19]. TFs regulating anthocyanin biosynthesis have also been identified in eggplant, such as SmMYB113, SmGL3[20−23]. In addition, anthocyanin production is affected by various environmental conditions including light and temperature[24−27]. Among them, light is a particular important regulator of anthocyanin biosynthesis[28−31]. However, the anthocyanins-related pigmenting is not or less influenced under darkness in specific materials of sweet cherry, chrysanthemum, mango, turnip, grape, and eggplant[20,32−36].
Fruit color under calyx is an indicator of light requirement in eggplant[37,38]. Specifically, fruits with green peel under the calyx always undergo a color change to white after bagging, which is known as the photosensitive or light-induced type[39,40]. In contrast, fruits with purple peel under the calyx often maintain their purple coloration after bagging, and these eggplants are referred to as the non-photosensitive type[20]. Most quantitative trait locus (QTLs) responsible for purple coloration under calyx in eggplants were detected on chromosome 10[41−45]. In addition, a recent study showed that SmFTSH10 (filamentation temperature sensitive 10) was the most possible candidate gene of non-photosensitivity in eggplant[45]. Interestingly, we found that some cultivars exhibited light purple under the calyx, while under bagging conditions, the fruits displayed a purple coloration overall, which was significantly lighter compared to the fruits that grew under natural conditions. Therefore, we classify these varieties as the less-photosensitive type.
In this study, two eggplant parental lines with photosensitive and less-photosensitive coloration and their crossing F2 progenies were used for genetic analysis, bulked sergeant analysis (BSA)-based sequencing and expression analysis to identify the causative genes conferring less-photosensitive anthocyanin biosynthesis. The candidate genes identified in this study would facilitate gene identification for the less-photosensitivity trait and be helpful for the study of the mechanism of less-photosensitive anthocyanin biosynthesis in eggplant.
-
A photosensitive cultivar '749' and a less-photosensitive cultivar '609' were used as parent lines to develop the F2 population. All eggplants grew in the greenhouse in the experimental fields of Hebei Agricultural University, Baoding, China. Eggplant fruits were bagged on the 5th day after flowering, and fruit color observation were conducted on the 14th day under bagging condition, with the fruits growing in natural conditions as control.
Whole genome sequence of bulked DNA
-
Total genomic DNA of the two parent lines and the F2 population was extracted from young leaves using a modified cetyltrimethylammonium bromide (CTAB) method[46]. For genome sequencing, equivalent amounts of DNA from 30 plants with purple-bagged and white-bagged fruits were mixed separately, named 'P Bulk' and 'G bulk' respectively. The qualified DNA was randomly broken into fragments with a length of 350 bp, and sequencing libraries were generated using a TruSeq Nano DNA HT Sample preparation Kit (Illumina USA), followed by Illumina PE150 sequencing.
After quality control, the clean reads of each sample were aligned against the eggplant reference genome[47] using BWA software[48]. The Unified Genotyper function in GATK3.8 software[49] was used to detect SNP and Indel of each sample, and Variant Filtration parameter in GATK was used to filter the SNPs and Indels. Euclidean Distance (ED) algorithm was employed to predicted candidate region associate with less-photosensitivity[50]. Sequencing depth of differential SNPs in each mixed pools were counted to calculate ED on each site. ED was raised to ED^5 to minimize noise of small variation[50].
Obtaining recombinants
-
To narrow down the candidate region, molecular markers (Supplemental Table S1) were developed based on the SNP sites of the two bulks from the sequencing. All plants in F2 population and the two parental lines were genotyped using Kompetitive allele specific PCR (KASP) technology with linked SNP markers, that were used to screen recombinants.
Gene expression analysis
-
Eggplant fruits of '609' and '749' were bagged on the 5th day after flowering, and the fruit peel was harvested on the 14th day after bagging for gene expression analysis, using the peel of fruits grown under natural conditions as control. The expression of candidate genes was detected using qRT-PCR. Primer Premier 5.0 software was used to design the primers, which are listed in Supplemental Table S2. Total RNA of fruit peel was extracted with RNAprep Pure Plant Plus Kit (Tiangen, Beijing, China). A total of 1 μg RNA per sample was reverse transcribed to cDNA using the PrimeScript™ reagent Kit with gDNA Eraser (TaKaRa, Beijing, China) in 20 μL of reaction mixture. The qRT-PCR was performed using THUNDERBIRD SYBR qPCR Mix (TOYOBO, Shanghai, China) in a LightCycler® 96 System (Roche, Basel, Switzerland). The expression of the candidate genes was quantified by 2−ΔCᴛ method, with SmGAPDH (EGP1067575) as housekeeping gene. The analysis was performed with three biological replicates. Significant differences between groups were assessed by one-way analysis of variance (ANOVA) followed by Tukey's test (p < 0.05) using SPSS 16.0 Statistics (SPSS Inc., Chicago, IL, USA).
-
Under natural growth conditions, both '609' and '749' exhibited dark purple fruit coloration, with the peel under calyx of '609' being light purple and that of '749' being green (Fig. 1). To investigate their responses to darkness, fruits were bagged to block the exposure to light. Under such conditions, '609' plants had significant lighter purple fruits compared to those grown under natural conditions, while '749' plants had white fruits (Fig. 1).

Figure 1.
Fruits of '609' and '749' under (L) natural and (D) bagging conditions. CUC indicates the color under calyx. Eggplant fruits were bagged on the 5th day after flowering, and the picture was taken on the 14th day after bagging, with the fruits growing in natural conditions as control.
The F1 hybrids ('609' × '749') and F2 populations were used for studying the inheritance of less-photosensitive trait. The fruit color after bagging of each plant was visually examined. It was shown that under bagging conditions, the fruits of 15 F1 individuals generated by crossing '609' and '749' displayed a light purple coloration. Among 178 F2 individuals, 139 and 39 plants had light purple and white fruits after bagging, respectively, and this rate approximately fitted an expected Mendelian inheritance ratio of 3:1 (χ2 = 0.91 < χ2 0.05,1 = 3.84) (Table 1). These results indicated that the less-photosensitive trait was controlled by a dominant gene, which was named as SmLP hereafter.
Table 1. Genetic analysis of fruit peel pigmentation after bagging in F2 population.
Generation Numbers of plants Number of plants with light purple fruit peel after bagging Number of plants with white fruit peel after bagging Expected ratio χ2 609 10 10 0 749 10 0 10 (609 × 749) F1 15 15 0 (609 × 749) F2 178 139 39 3:1 0.91 Note: χ2 0.05 = 3.84, df = 1. Identification of candidate region for SmLP through BSA analysis
-
To map SmLP, BSA-based sequencing was carried out by bulking 30 F2 individuals with white-bagged fruits and purple-bagged fruits, respectively. The high-throughput sequencing generated 78.25 Gb clean data, which comprised 122,455,776 and 138,367,408 high quality reads from 'G bulk' and 'P bulk'. The Q30 ratio was higher than 93.50% (Supplemental Table S3). The mapping rate exhibited an average cover depth of 99.63% (Supplemental Table S4). These results suggested that the sequencing data were reliable and suitable for SNPs and Indels detection.
A total of 60,648 poly morphic sites (43,725 SNPs and 16,923 Indels) were detected in the BSA data. The median plus three standard deviations (SD) of the fitted values at all loci were used as the association threshold for analysis, which was determined to be 0.49. Based on this association threshold, significant associations were detected on Chromosome 10 (Fig. 2a), spanning a total length of 15.82 Mb and located at 4.38−20.15 Mb (Fig. 2b).
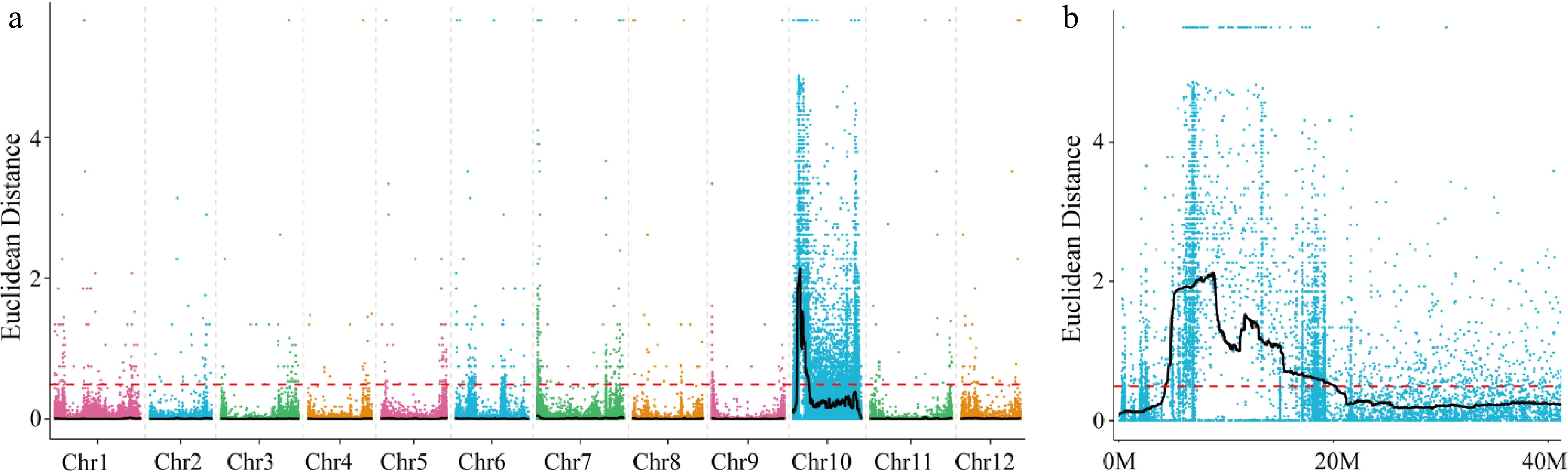
Figure 2.
Distribution of ED-based linkage value on (a) all chromosomes and (b) on Chromosome 10. Each colored dot represents an ED-based linkage value of an SNP site. Black lines represents ED value after fitting. Red dashed lines represents linkage threshold.
Further mapping of SmLP by screening recombinants
-
In order to further determine the positioning range of SmLP, 178 individuals in the F2 population, and 10 KASP primers, which were designed to uniformly cover the preliminary mapping interval, were used to analyze the polymorphism of the two parental lines. The SmLP locus was finally mapped to a region between the markers SNP9 and SNP3 (with a physical ranging from 7.4 Mb to 12.5 Mb), based on 52 recombinant individuals (Fig. 3). Within the candidate interval, there were 280 SNPs and 74 Indels in total (Table 2). These SNPs and Indels were associated with 116 genes (Supplemental Table S5), among which six genes had non-synonymous mutations (Table 3).
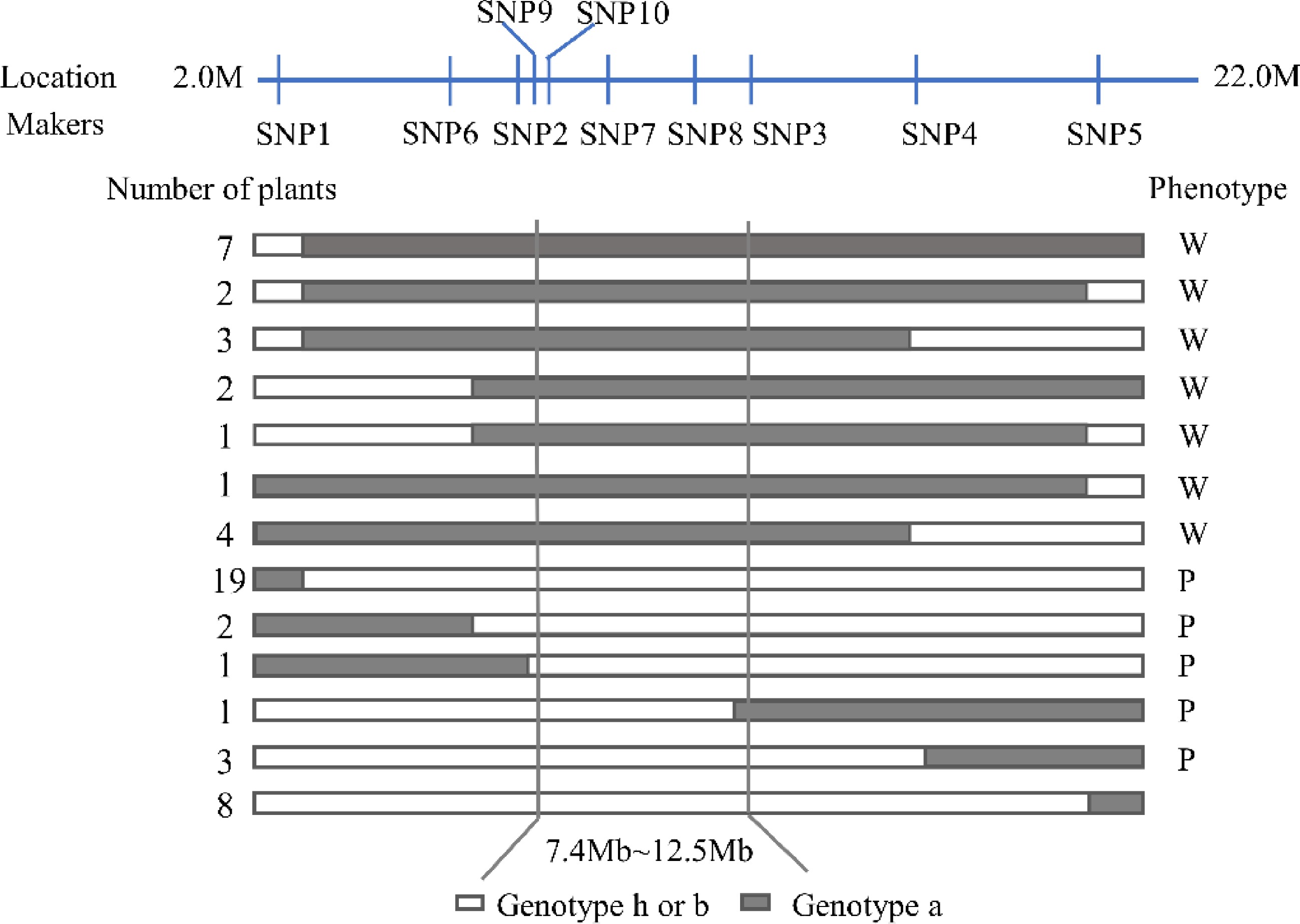
Figure 3.
Genotype and phenotype analysis of recombinant plants in the F2 population derived from a cross between '609' and '749'. (a) Genotype of the photosensitive parent '749'. (b) Genotype of the less-photosensitive parent '609'. (h) Heterozygote of the '749' and '609'. W, White fruit peel after bagging; P, Purple fruit peel after bagging.
Table 2. Classifications of SNPs and Indels in the candidate region.
Category The number of SNPs The number of Indels Intergenic 222 56 Upstream 23 5 3'UTR 0 1 Non-synonymous 6 0 Synonymous 1 0 Intronic 9 1 Downstream 19 11 Table 3. Nonsynonymous SNPs and their related genes in the candidate region.
Gene ID SNP loci Base substitution type Annotation EGP21857 7447780 C- > T Uncharacterized protein LOC102595296 EGP21873 7722116 C- > G 12-oxophytodienoate reductase 1 EGP21911 9870254 C- > T Undefined EGP21972 11985911 C- > T Hypothetical protein BC332_00197 EGP21983 12225282 G- > A Putative GDSL esterase/lipase-like EGP22005 12532757 G- > A MYB domain protein 113 Identification of candidate genes potentially regulating less-photosensitivity
-
The biosynthesis pathway of anthocyanin has been studied and characterized very clearly in different plants; related genes controlling the pathway have been classified into structural genes, encoding enzymes that directly catalyze stepwise the anthocyanin biosynthesis process, and regulatory genes controlling the expression of structural genes[13]. The expressions of structural genes and regulatory genes were influenced by both intrinsic biological factors (such as hormones, circadian rhythms) and external environmental factors (such as light, temperature, insects, and fungi)[25,30,51−57]. Based on gene functional annotation and homologous gene functional studies, the SmLP region was analyzed in search of anthocyanin biosynthesis-related and light signal transduction-related genes, and there were seven high confidence genes observed in this region (Table 4). Among them, there were three MYB transcription factors. Both EGP21874 and EGP21875 encode a TF SmMYB113, and they were homologs of AtMYB113 in Arabidopsis. AtMYB113 was identified as one of the members of the MBW complex, directly regulating the expression of structural genes[58]. EGP22005 is homologs of the gene AtMYB16 in Arabidopsis, which is involved in controlling trichome maturation and cuticle formation[59,60]. In addition, EGP21863 encodes auxin response factor 16 (ARF16), and some ARFs were known to negatively regulate the biosynthesis of anthocyanins, such as ARF13 and ARF2[61,62]. EGP21864 encodes a phototropic-responsive NPH3 family protein. EGP21891 and EGP21908 encode phytochrome kinase substrates (PKSs). It was reported that under blue light, phototropins (PHOTs) could interact with NPH3 and PKSs, regulating the bending of the hypocotyl during phototropism[63−65]. Therefore, EGP21864, EGP21891, and EGP21908 may be involved in responses to light, particularly to blue light.
Table 4. Candidate genes involved in biosynthesis of anthocyanin and light signal transduction.
Gene ID SNP category Indel category Annotation EGP21874 Intergenic region in upstream, Intergenic region in downstream Intergenic region in downstream MYB domain protein 113 EGP21875 Intergenic region in upstream, Intergenic region in downstream − MYB domain protein 113 EGP22005 Upstream, Non-synonymous − MYB domain protein 16 EGP21863 Upstream Intergenic region in upstream Auxin response factor 16 EGP21864 Intron − Phototropic-responsive NPH3 family protein EGP21891 Intron, Downstream, Intergenic region in downstream Intergenic region in downstream Phytochrome kinase substrate 2 EGP21908 Intergenic region in downstream Intergenic region in downstream Phytochrome kinase substrate 1 The expression analysis of seven genes potentially participating anthocyanin biosynthesis and light signal transduction (Table 4) and six genes with non-synonymous mutations (Table 3) showed that only EGP21875, EGP21864, and EGP21911 were expressed in the '609' and '749' peel (Fig. 4, Supplemental Fig. S1). Therefore, these three genes were considered as putative genes controlling less-photosensitive coloration in eggplant.
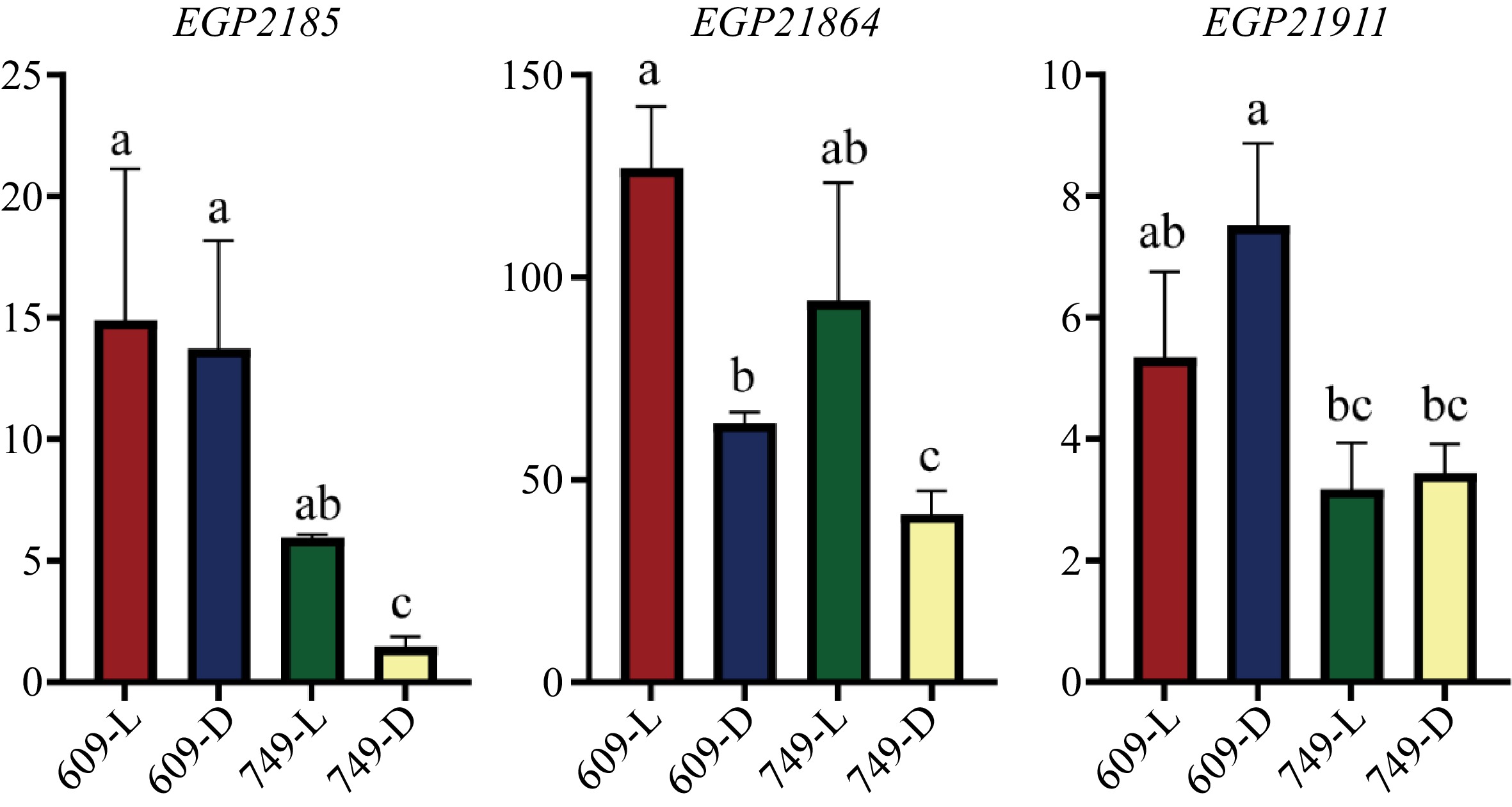
Figure 4.
The transcript level of EGP21875, EGP21864 and EGP21911 in the fruit peel of '609' and '749'. The y axis indicated the relative expression levels of each gene. L, Under natural conditions; D, Under bagging conditions. The relative expression was determined by 2−ΔCᴛ method. The date are means from three biological replicates with three technical replicates. Error bars indicate SEs. Letters above each column represent significant differences based on one-way analysis of variance (ANOVA) followed by Tukey's test (p < 0.05).
EGP21875 encodes a MYB TF SmMYB113, which has been reported to participate in regulating the biosynthesis of anthocyanin in eggplant[20,22,23]. In this study, the expression of SmMYB113 in the '749' peel was significantly downregulated after bagging. Whereas the expression of SmMYB113 in the '609' peel was not affected by light (Fig. 4). There was one SNP 22.9 kb upstream of the start codon and one SNP 13 kb downstream of the stop codon of EGP21875, respectively (Table 5).
Table 5. Candidate genes involved in less-photosensitive anthocyanin biosynthesis in the peel of '609'.
Gene ID SNP loci Substitution type SNP category Distance Annotation EGP21875 7808145
7770881C- > A
T- > CIntergenic region in upstream, Intergenic region in downstream 22,893 bp
13,006 bpMYB domain protein 113 EGP21864 7625998 C- > T Intron − Phototropic-responsive NPH3 family protein EGP21911 9870254 C- > T Non-synonymous − − EGP21864 encodes a phototropic-responsive NPH3 (Non-Phototropic Hypocotyl 3) family protein. Studies have shown that AtNPH3 is involved in regulating photomorphogenesis and light-mediated growth responses in Arabidopsis[66,67]. After bagging, the expression of EGP21864 was downregulated in the peel of both '609' and '749'. Notably, under bagging conditions, the expression of EGP21864 in the peel of '609' was significantly higher than that in '749' (Fig. 4). There was one SNP within the intronic region of EGP21864 (Table 5).
EGP21911 encodes a protein with unknown function. A SNP (G/A) in '609' identified through sequencing analysis resulted in an amino acid change from Ala-15 to Thr-15 in the first exon (Table 5). Under natural conditions, EGP21911 showed no distinctively different expression level between the two parental lines. However, under bagging conditions, the expression of EGP21911 was significantly higher in the peel of '609' compared with '749' (Fig. 4).
-
Eggplant fruit pigmentation is usually affected by low light conditions[39]. While the accumulation of anthocyanins in less-and non-photosensitive varieties was less influenced by darkness (Fig. 1)[20,38], thus identification of the genes responsible for less- and non-photosensitivity is crucial for breeding low light-tolerant varieties. Most previous studies focused on mapping genes related to purple coloration under the calyx, but did not distinguish between less- and non-photosensitivity[41,43,44]. A related study reported that a dominant gene controlling non-photosensitivity in eggplant was mapped between 19.9−20.2 Mb on chromosome 10[45], which is close to, but does not overlap with the candidate region (7.4−12.5 Mb) we identified for controlling less-photosensitivity in this study. Considering the different mapping results and the distinct phenotypes observed after bagging treatment, we speculated that the loci controlling non-photosensitivity and less-photosensitivity in eggplant may be different.
In this study, we identified three putative genes conferring less-photosensitivity in eggplant. EGP21875 encodes a MYB TF SmMYB113, which plays critical roles in regulating anthocyanin biosynthesis by activating the expression of structural genes[22,23]. Overexpression of SmMYB113 in eggplant resulted in a significant upregulation of the expression levels of structural genes, such as SmCHS, SmCHI, SmF3H and SmANS, and a substantial accumulation of anthocyanins in the regenerating shoots of eggplant[23], indicating that upregulation of the SmMYB113 may directly increase anthocyanin accumulation. Moreover, the expression level of EGP21875 in the peel of '609' was significantly higher than that of '749' under bagging conditions (Fig. 4). Therefore, the coloration of the '609' fruit peel under bagging conditions may be associated with the upregulation of SmMYB113 expression, which may attribute to SNPs in the intergenic region.
Another potential candidate is EGP21864 encoding a phototropism response NPH3 family protein. Studies have shown NPH3 is involved in the perception and transduction of light signals, allowing plants to properly orient their growth towards light. The hypocotyl of nph3 mutant in Arabidopsis thaliana failed to exhibit bending towards light and remains mostly straight, resulting in a defective phototropic response[66−68]. The expression of EGP21864 was down-regulated both in the peel of '609' and '749' under bagging conditions, compared with that in natural conditions. And the expression of EGP21864 in '609' was significantly higher than that in '749' under bagging conditions (Fig. 4). Therefore, the SNP of EGP21864 may affect the sensitivity of plants to light. However, there was no literature or report indicating the direct involvement of phototropism response NPH3 family protein in anthocyanin biosynthesis, and future functional studies should be conducted to validate its specific contribution to fruit color determination.
In addition, EGP21911, a gene with non-synonymous mutation SNP, encodes a protein with unknown function. Its expression in '609' and '749' peel was not regulated by light. Under bagging condition, the expression in the peel of '609' was significantly higher than that of '749'. Whether this gene is involved in anthocyanin biosynthesis remains to be further studied.
This study focused on mining the genes responsible for less-photosensitivity using BSA analysis, while other reported studies have utilized various methods such as traditional QTL mapping and Genome Wide Association Studies (GWAS) to identify the locus for fruit color under the calyx[41−45]. In BSA analysis, individuals with similar phenotype are grouped together, and molecular markers are used to analyze the pooled samples in order to identify loci associated with the traits of interest. BSA analysis has been widely utilized in gene mapping due to its advantages of high efficiency, low cost, time-saving, and labor-saving[69−72]. QTL mapping has been proved to be a powerful method to identify regions of genome that co-segregate with a given trait, especially quantitative traits. But QTL mapping is highly time-consuming and labor-intensive[73−76]. Based on linkage disequilibrium, GWAS is an effective analytical tool for deciphering the genetic basis of phenotypic diversity in crops. It possesses significant advantages of high throughput, efficiency, and reduced time consumption[77−79]. The aforementioned studies were primarily conducted on the basis of second-generation sequencing or existing molecular markers, obtaining multiple QTLs, candidate genes, or molecular markers associated with fruit color under the calyx[41−45]. Additionally, except for SNPs and Indels, some tissue color variations in horticultural crops were reported to be related to variations in large genomic segments, such as promoter variations, insertion of transposons, multiple repeats sequence, deletion of segments, etc[80−86]. The detections of such variations may necessitate more refined sequencing techniques, such as third-generation sequencing.
In summary, EGP21875 encoding a MYB TF, EGP21864 encoding a phototropic response NPH3 family protein, and EGP21911 encoding an unknown functional protein were putative genes for regulating less-photosensitive anthocyanin biosynthesis in eggplant. Of note, EGP21875 (SmMYB113) was the best candidate gene. Future studies, including functional validation and genetic mapping, will help to unravel the intricate regulatory networks and molecular mechanisms involved in less-photosensitive coloration in eggplant.
-
The less-photosensitive pigmentation in eggplant was controlled by a single dominant gene. The causal gene was mapped on chromosome 10, spanning from 7.4 Mb to 12.5 Mb. Three candidate genes, namely EGP21875 (MYB domain protein 113), EGP21864 (Phototropic-responsive NPH3 family protein) and EGP21911 (Unknown protein), were identified as the putative genes conferring less-photosensitive coloration in eggplant.
-
The authors confirm contribution to the paper as follows: conceptualization, investigation, visualization, writing—original draft, writing—review & editing: Luo L; methodology, software, investigation: Niu Y; conceptualization, visualization, writing—review & editing: Li Q; methodology, software: Xia L, Wang C; investigation, data curation: Luo S; writing—review & editing, visualization: Li N, Xuan S, Wang Y; supervision, funding acquisition: Shen S; writing—review & editing, supervision, funding acquisition: Zhao J, Chen X; data curation: Chen X. All authors reviewed the results and approved the final version of the manuscript.
-
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
This work was supported by grants from Hebei Province Innovative Team Construction Project for the Third Phase of Modern Agricultural Industry Technology System (HBCT2023100207), International Joint R & D Center of Hebei Province in Modern Agricultural Biotechnology, and the Post-graduate's Innovation Fund Project of Hebei (CXZZBS2019098). Finally, we gratefully acknowledge the help of Yiguo Hong from Hebei Agricultural University College of Horticulture in revising this article.
-
The authors declare that they have no conflict of interest. Jianjun Zhao is the Editorial Board member of Vegetable Research who was blinded from reviewing or making decisions on the manuscript. The article was subject to the journal's standard procedures, with peer-review handled independently of this Editorial Board member and the research groups.
- Supplemental Table S1 Primer sequence for KASP detection.
- Supplemental Table S2 List of primer used for RT-PCR analysis.
- Supplemental Table S3 Summary of sequencing data quality.
- Supplemental Table S4 Summary of sequencing depth and coverage statistics.
- Supplemental Table S5 Genes in candidate region related to less photosensitive anthocyanin biosynthesis of '609'.
- Supplemental Fig. S1 The transcript level of candidate genes with low expression in the fruit peel of '609' and '749'.
- Copyright: © 2023 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Luo L, Niu Y, Li Q, Xia L, Wang C, et al. 2023. Mapping and identification of genes responsible for less-photosensitive fruit coloration in eggplant. Vegetable Research 3:32 doi: 10.48130/VR-2023-0032
Mapping and identification of genes responsible for less-photosensitive fruit coloration in eggplant
- Received: 30 September 2023
- Accepted: 31 October 2023
- Published online: 21 December 2023
Abstract: In eggplant (Solanum melongena. L), low light during cultivation often hinders proper pigmentation of fruit. While some varieties exhibit less susceptibility to low light for eggplant coloration, however, the genetic basis of such less-photosensitive fruit coloration remains unknown. In this study, we characterized a less-photosensitive eggplant cultivar '609'. Under bagging conditions, fruits of '609' exhibited purple coloration, albeit lighter than fruits grown under natural conditions. Genetic analysis showed that the less-photosensitive trait was controlled by a single dominant gene, designated SmLP. Based on BSA and genetic recombination analyses, SmLP was mapped to the 7.4−12.5 Mb region on chromosome 10. Within this genetic region, six genes with non-synonymous mutation and seven genes potentially involved in anthocyanin biosynthesis or light signal transduction were identified. Further RT-qPCR analysis revealed that only three out of these genes were differentially expressed in eggplant peel tissues. The three genes EGP21875, EGP21864 and EGP21911, encoding MYB domain protein 113, phototropic-responsive NPH3 family protein, and protein with unknown function, respectively, were considered as the putative genes associated with less-photosensitive anthocyanin biosynthesis in eggplant fruits. These results would provide great help in casual gene identification for less-photosensitive trait and promote an understanding of molecular mechanisms underlying less- and non-photosensitive coloration in eggplant.
-
Key words:
- Eggplant /
- Anthocyanin /
- Less-photosensitive /
- BSA













