-
RNA editing is an epitranscriptomic process that modifies genetic information directly at the RNA level, involving nucleotide insertion, deletion, or substitution[1]. Among the various types of RNA editing, adenosine-to-inosine (A-to-I) editing is particularly prevalent in nuclear-encoded messenger RNA (mRNA). This editing was first discovered in the oocytes and embryos of Xenopus laevis in 1987[2] and has since been found to be conserved across metazoans[3,4]. In 2016, A-to-I mRNA editing was identified in the fungal plant pathogen Fusarium graminearum[5], significantly broadening our understanding of this process. Further studies revealed that A-to-I editing is common within the Sordariomycetes class of fungi[6−8]. In 2017, it was also reported in bacteria[9]; however, unlike in animals and fungi, only a limited number of A-to-I editing sites have been identified in the studied bacterial species[9−11].
In animals, A-to-I RNA editing is mediated by the Adenosine Deaminase Acting on RNA (ADAR) enzyme family[12], a metazoan-specific innovation that originated in the last common ancestor of existing metazoans[4]. ADAR proteins possess a conserved domain architecture, featuring multiple dsRNA binding domains (dsRBDs) at the N-terminus and a catalytic deaminase domain at the C-terminus[13]. Notably, ADAR genes are preferentially expressed in the nervous system[14,15]. In bacteria, A-to-I mRNA editing is facilitated by tRNA-specific adenosine deaminase A (TadA), known for editing A34 in tRNAArg[16,17]. In fungi, the A-to-I mRNA editing machinery consists of Tad2, Tad3, and Ame1[18]. The Tad2-Tad3 complex is a conserved heterodimeric deaminase responsible for editing the wobble position (A34) in 7−8 cytosolic tRNAs in eukaryotes[16]. Ame1, a sexual stage-specific cofactor that originated in the last common ancestor of Sordariomycetes, enables the Tad2-Tad3 complex to edit mRNA specifically during sexual reproduction by interacting with the N-terminal domain of Tad3[18].
Inosine, when recognized by cellular machinery, is interpreted as guanosine. Consequently, A-to-I RNA editing effectively functions as an A-to-G mutation at the RNA level, potentially leading to protein recoding. While metazoans exhibit abundant A-to-I RNA editing sites, recoding (nonsynonymous) editing sites typically constitute a minor fraction[3,4]. The majority of A-to-I editing sites occur in noncoding regions, often within repetitive elements. Coleoid cephalopods are a notable exception, showcasing an average of 60,000 recoding editing sites per species, affecting nearly half of their protein-coding genes[19]. Drosophila also exhibit a relative enrichment of recoding editing sites despite having fewer total A-to-I editing sites compared to most other animals[20,21]. Sordariomycetes, on the other hand, exhibit tens of thousands of editing sites, with most resulting in protein recoding[5,8,22,23]. Even in coleoid cephalopods, recoding editing remains a small fraction of all editing sites (11%–13%)[4,24]. Despite being rare, RNA editing in bacteria often results in protein recoding[9−11].
In this review, we summarize recent findings on A-to-I RNA editing across animals, fungi, and bacteria, with a focus on the functional roles and adaptive advantages of recoding editing. We propose a universal adaptive role for A-to-I recoding editing in resolving evolutionary trade-offs, emphasizing how this mechanism addresses conflicting demands, such as survival and reproduction. Additionally, we discuss current evolutionary analyses and experimental methods used to determine the function and adaptive advantages of recoding editing, identifying their limitations. Finally, we propose improvement strategies to address these shortcomings, aiming to advance our understanding of the evolutionary and functional significance of A-to-I recoding editing.
-
Not all adenosines in RNA substrates are edited. In animals, ADAR-mediated A-to-I editing preferentially occurs on dsRNA. Two main types of editing are identified[25]: selective editing, which targets one or a few specific adenosines within imperfect dsRNA structures in the coding region of mRNA, and nonselective editing (hyper-editing), which involves multiple adenosines within perfectly or nearly perfectly matched long dsRNA, often found in noncoding regions and repetitive elements. Many editing sites in protein-coding regions are located within repetitive elements, resulting from hyper-editing rather than selective editing. For selective editing, an imperfect fold-back dsRNA structure forms between the exon sequence around the editing site and a downstream complementary sequence called the editing-site complementary sequence (ECS)[26]. Multiple RNA sequences and structural features have been proposed to influence ADAR editing[25,27]. While ADARs do not follow a strict consensus sequence, they show weak nucleotide preferences near the editing site: a bias against G at the −1 position and slight enrichment for G or A at the +1 position[3].
Given that A-to-I mRNA editing in both fungi and bacteria is mediated by A34 tRNA editing enzymes, it is not surprising that the editing sites in these organisms show a preference for hairpin loop structures similar to the anticodon loop structures of tRNA, rather than dsRNA structures[9−11,17,18,22]. Furthermore, A-to-I mRNA editing sites in these organisms also exhibit similar base preferences at adjacent positions to tRNA editing, particularly a preference for uracil (U) at the −1 position[9−11,17,18,22]. In fungi, while hairpin loop structures contribute significantly to editing, primary sequences have a pronounced influence on editing[22]. This is particularly evident with the critical role of the U at the −1 position for editing in fungi.
-
Unlike genomic mutations, which uniformly affect all mRNAs transcribed from a gene, RNA editing often occurs partially, with an editing level—the percentage of edited transcripts relative to total transcripts at a given site—ranging between 0 and 100%. This binary A/I code allows for the simultaneous expression of both edited and unedited isoforms from a single gene. Moreover, editing levels can vary across tissues and developmental stages[8,28], providing organisms with a dynamic mechanism to regulate gene expression by spatially and temporally adjusting the proportion of edited and unedited isoforms. Consequently, RNA editing enhances transcript/protein diversity and functional flexibility, making it particularly advantageous for resolving trade-offs caused by pleiotropy across different tissues or stages. Recent genetic studies in fungi have demonstrated that sexual stage-specific A-to-I editing can address survival-reproduction trade-offs caused by genes that are essential for sexual reproduction but harmful to survival during vegetative growth[29].
Additionally, the continued expression of unedited isoforms means that recoding editing incurs a low evolutionary cost. This allows for the generation of amino acid residues that cannot be achieved through direct mutations in the genomic sequence. Although an edited A may not inherently confer more advantages than a genomically encoded G, A-to-I recoding can still provide adaptive value in the absence of beneficial A-to-G mutations at this site. Thus, recoding editing may act as a transitional mechanism, potentially facilitating the eventual fixation of advantageous genomic A-to-G mutations[30−32].
-
If a recoding event confers an adaptive advantage, it is likely to become fixed in the population through positive selection and maintained by purifying selection. Context-dependent genomic mutations can further optimize the editing level at these sites by fine-tuning the surrounding sequence context. Comparisons of synonymous (assumed neutral) and nonsynonymous editing frequencies have been used to infer the adaptive nature of recoding. In humans, these comparisons suggest that recoding editing is generally deleterious, with lower frequencies and levels of nonsynonymous editing compared to synonymous editing[33]. Similarly, coleoid cephalopods and Drosophila also exhibit lower frequencies and levels of nonsynonymous editing compared to synonymous editing across identified recoding sites[20,21,34,35]. However, analysis of conserved editing across multiple species reveals a higher frequency of conserved nonsynonymous editing sites compared to conserved synonymous editing sites in humans, mice, coleoid cephalopods, and Drosophila[20,21,34−36]. In contrast, fungi exhibit higher frequencies and levels of nonsynonymous editing compared to synonymous editing, even among the total identified recoding sites[8,37]. These results suggest that while only a subset of recoding sites (i.e., conserved) are likely adaptive in animals, recoding sites in fungi are generally adaptive.
-
Examining the maintenance of recoding events through purifying selection can also help determine the adaptive nature of recoding editing. A recoding site can be classified as neutral or slightly deleterious (A-preferring), primarily a transitional state (G-preferring), or a means to increase protein diversity and flexibility (A/I-preferring) (Fig. 1a). G-preferring recoding sites are expected to have a genomic A-to-G substitution rate during evolution that is higher than that at synonymous editing sites (Fig. 1b). In contrast, A-preferring recoding sites should exhibit an A-to-G substitution rate that is equal to or lower than that at synonymous editing sites, but not less than that at nonsynonymous unedited counterparts (Fig. 1b). Since these sites accommodate editing, having a G at these positions is likely less harmful than at typical nonsynonymous unedited sites[33]. The rate of A-to-G substitutions at A/I-preferring sites is anticipated to be lower compared to nonsynonymous unedited counterparts (Fig. 1b), as such substitutions could result in a loss of the beneficial traits associated with increased protein diversity and flexibility.

Figure 1.
Classification of A-to-I recoding sites and their evolutionary dynamics. (a) Recoding sites can be classified as A-preferring, G-preferring, and A/I-preferring based on the fitness effects of the edited and unedited variants. Editing at A-preferring sites is neutral or slightly deleterious, becoming fixed in the genome primarily through genetic drift. A/I-preferring sites are positively selected for their ability to enhance protein diversity and flexibility. In contrast, G-preferring sites may be fixed through positive selection due to their compensatory or transitional roles, or through genetic drift if they are harm-permitted by editing. These sites can also be categorized into restorative or diversifying editing based on the amino acid changes relative to ancestral states. Restorative editing includes compensatory editing, which corrects pre-existing harmful genomic G-to-A mutations, and harm-permitting editing, which corrects genomic G-to-A mutations that it itself promotes. Both G-preferring and A/I-preferring compensatory editing are adaptive, while only A/I-preferring harm-permitting editing can be adaptive. (b) The expected A-to-G substitution rate at the three types of recoding sites is shown relative to synonymous editing sites (neutral rate) and nonsynonymous uneditable sites.
Estimating the maintenance of recoding events through phylogeny and ancestral sequence reconstruction suggests that recoding editing in Drosophila and conserved recoding editing in the coleoid lineage are less likely to serve as transitional states for G-preferring substitutions. In the coleoid lineage, a significantly lower frequency of A-to-G substitutions at highly conserved recoding sites has been observed compared to synonymous editing sites[19,38]. A similar pattern is evident in Drosophila, where recoding sites also show a significantly lower frequency of A-to-G substitutions than synonymous editing sites[39]. However, this estimation method is only applicable to sites shared by at least two species, making it incapable for determining the adaptive nature of newly generated or non-conserved editing sites. It is important to note that A-preferring recoding edits tend to be eliminated while G-preferring edits can become genomically encoded over evolutionary time. Consequently, we expect these two types of edits to be less frequent in conserved editing sites shared across species and more common in newly generated sites. Indeed, population genomics approaches have shown that edited A nucleotides are more frequently substituted with G compared to their unedited counterparts in coleoid cephalopods, Drosophila, and humans[30,31].
Additionally, to conclusively determine whether these conserved recoding sites are A/I-preferring, it is crucial to compare their A-to-G substitution rates to those of nonsynonymous unedited counterparts rather than solely to synonymous editing sites (Fig. 1b). In humans, nonsynonymous edited A nucleotides are more likely to be replaced with G or T/C compared to unedited A nucleotides[33]. In contrast, fungi exhibit the opposite trend[8], indicating that recoding editing in fungi offers an adaptive advantage for protein diversity and flexibility, leading to the maintenance of these recoding events by natural selection during evolution.
-
The presence of editing activity can relax functional constraints at the genomic level, allowing G-to-A mutations to create new edited A sites[40], resulting in restorative editing. Recently, a harm-permitting model[38] proposed that restorative editing is non-adaptive because the derived genotype with a highly edited A does not confer a fitness advantage over the original genomic G[38]. Researchers categorized nonsynonymous editing sites in coleoid species into restorative editing, which reverts the amino acid to its ancestral state, and diversifying editing, which changes the amino acid to a non-ancestral state[19,38]. They found a higher frequency and level of strongly edited (editing level > 10%) restorative editing sites compared to synonymous strong sites, but not for diversifying editing sites[19,38]. This led them to conclude that the observed excess of nonsynonymous editing in coleoid cephalopods is explained by a harm-permitting model and is nonadaptive[38]. However, if restorative editing is non-adaptive and merely harm-permitting, G-to-A mutations should be fixed by genetic drift, with a frequency no higher than neutral mutations. The observed higher frequency of restorative editing compared to synonymous editing sites strongly suggests that positive selection promotes the initial fixation of G-to-A mutations that lead to restorative editing, or that purifying selection prevents the loss of beneficial restorative editing. This indicates adaptive restorative editing. Contrary to previous claims[38], the harm-permitting model predicts only a higher median level of editing, not a greater frequency of restorative editing relative to synonymous editing.
It is important to conceptually distinguish between two types of restorative editing that address different sources of genomic mutations: compensatory and harm-permitting (Figs 1a & 2). Compensatory editing corrects pre-existing genomic mutations, aligning with the traditional concept of restorative editing[41]. If the genomic mutation is harmful, compensatory editing is considered adaptive, as it offsets the negative effects of the mutation. This type of editing can be viewed as a transitional state. If its primary function is to correct deleterious G-to-A mutations, we would expect to observe more frequent A-to-G substitutions at these sites, given that editing levels are typically below 100%. Although compensatory editing is expected to be less common due to purifying selection eliminating deleterious mutations before RNA editing emerges[40], it can still occur when genomic mutations that were once advantageous and became fixed in a population have now turned harmful. In contrast, harm-permitting editing corrects genomic mutations that it itself promotes. Merely mitigating the negative effects of genomic mutations does not necessarily imply that harm-permitting editing is adaptive.
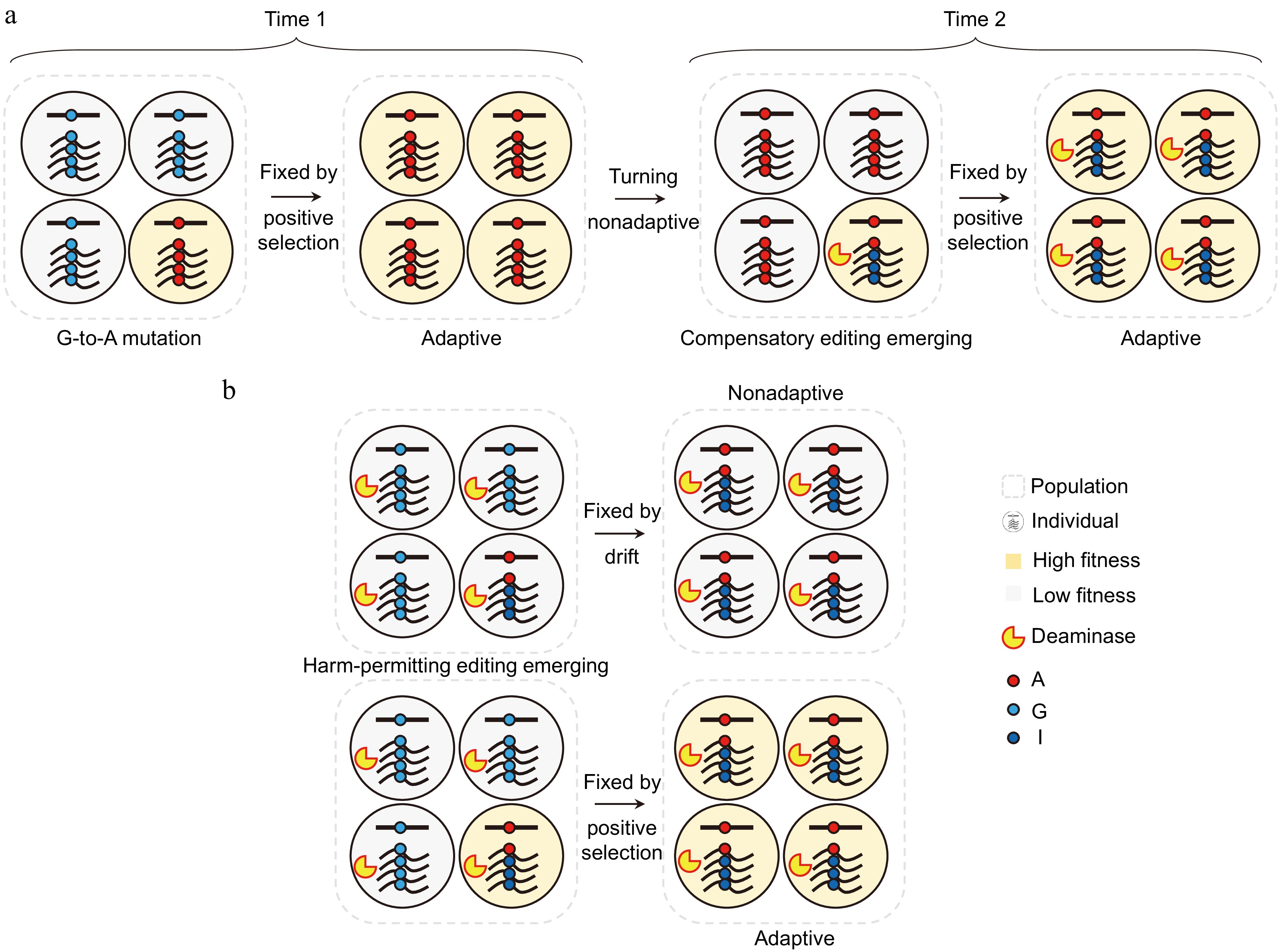
Figure 2.
The potential evolutionary processes of two types of restorative editing: compensatory editing and harm-permitting editing. (a) When G-to-A mutations that were once advantageous at Time 1 become harmful at Time 2, compensatory A-to-I editing can be established through positive selection due to its role in compensating for these mutations or enhancing protein diversity and flexibility. (b) Pre-existing editing activity can relax functional constraints at the genomic level, allowing G-to-A mutations to create new edited A sites, resulting in harm-permitting editing. This process underscores the intricate relationship between A-to-I editing and G-to-A mutations, as the formation of edited A sites relies on A-to-I editing. Consequently, no intermediate individuals can possess the corresponding editable A sites without the associated editing events. If harm-permitting editing only serves a restorative function, it should be nonadaptive and fixed by genetic drift, as it does not confer a fitness advantage over the original genomic G. However, it can be adaptive and fixed by positive selection when the protein diversity and flexibility provided by harm-permitting editing confer a competitive advantage.
Categorizing RNA editing as either restorative or diversifying based solely on amino acid changes at the editing site does not fully capture its unique potential for enhancing protein diversity and flexibility (A/I-preferring). In addition to its restorative function, the protein diversity and flexibility provided by restorative recoding may also contribute to its adaptive significance. Recent experimental studies in fungi highlight this distinction, showing that a flexibly edited A can confer higher fitness than an uneditable A or the original genomic G, regardless of whether the editing is classified as restorative or diversifying[29,42]. Furthermore, the definition of diversifying editing in this context differs from the traditional diversifying hypothesis of RNA editing, which suggests that it generates multiple variants from a single gene either simultaneously or spatially due to the binary A/I code and combinations of multiple editing sites[15,26,43].
The significantly lower frequency of diversifying compared to synonymous editing in each coleoid species examined strongly suggests that diversifying editing is generally deleterious and has been selectively purged[38]. Only diversifying editing shared by different coleoid cephalopods shows adaptive signals, indicating that a fraction of evolutionary conserved sites are adaptive[38]. In contrast, fungi exhibit higher frequencies and levels of both restorative and diversifying editing compared to synonymous editing sites (our unpublished data), strongly suggesting that both types of editing in fungi is generally adaptive.
-
While evolutionary analysis can suggest whether recoding editing is adaptive, it remains largely unknown which specific recoding sites are functionally important and adaptive, and what advantages these editing events provide over genomic mutations. Although challenging and time-consuming, experimental studies on the functional consequences of individual recoding sites are essential for addressing these fundamental questions. For example, recent genetic research in fungi has shown that sexual stage-specific A-to-I editing activity promotes G-to-A nonsense mutations in genes crucial for sexual reproduction but detrimental to survival[29]. This leads to harm-permitting restorative editing that addresses survival-reproduction trade-offs, helping us understand the adaptive advantage of this type of editing (Fig. 3).
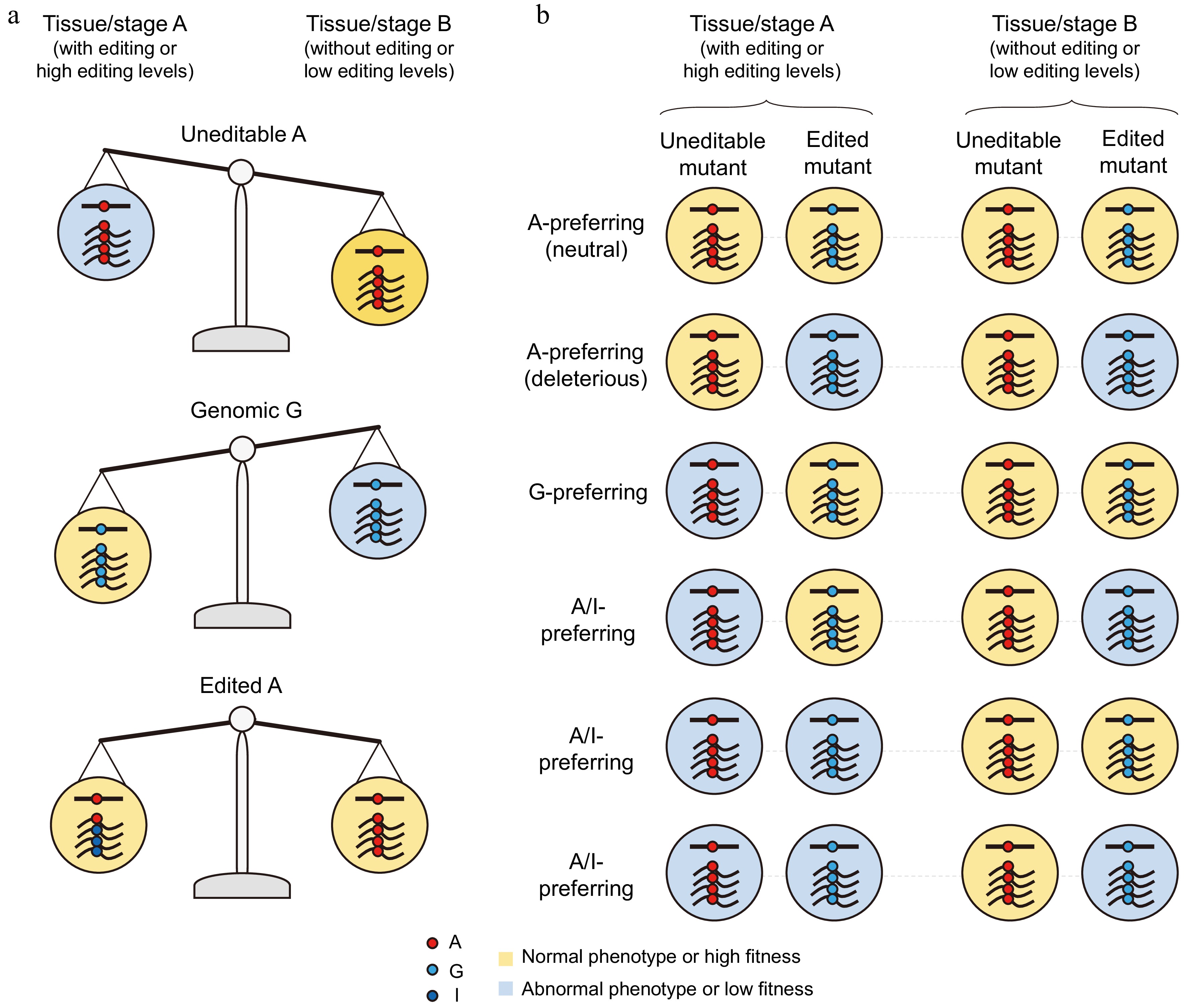
Figure 3.
The advantages of A-to-I RNA editing in navigating trade-offs arising from antagonistic pleiotropy and the anticipated phenotypes or fitness of uneditable and edited mutants. (a) The A variant is detrimental in tissue/stage A but beneficial in tissue/stage B, and/or the G variant is beneficial in tissue/stage A but detrimental in tissue/stage B. Tissue/stage A-specific RNA editing can alleviate the trade-offs resulting from the antagonistic pleiotropy of these two variants across both tissues/stages. (b) The anticipated developmental or physiological phenotypes and fitness of uneditable mutants (expressing only the A variant) and edited mutants (expressing only the G variant) for each type of recoding sites, as determined through genetic studies.
The gold standard for assessing the function of individual recoding sites is to create genetic mutants that prevent or reduce editing at target sites without altering the encoded amino acid sequences (Fig. 3). Observing developmental or physiological defects in these mutants in tissues or stages where editing occurs would indicate that the recoding event is functionally important for the organism. However, generating uneditable or editing-impeding mutants is not straightforward. Among the 64 genetic codons, only the nonsynonymous edited codons for Isoleucine (I) (ATA), Arginine (R) (AGA/AGG), and Serine (S) (AGT/AGC) can be directly replaced with their uneditable synonymous counterparts (ATT/ATC for I, CGT/CGC/CGA/CGG for R, and TCT/TCC/TCA/TCG for S). In fungi and bacteria, this can be achieved by mutating nucleotides at the third position of the codons adjacent to the edited sites, given the strict sequence preferences of their editing enzymes. In animals, editing can be impeded by removing the essential ECS, but this is only feasible when the ECS is located within intron regions.
To further explore the adaptive advantages of functionally important recoding sites, we need to create edited mutants that express only the edited version of proteins by directly replacing the A with G at the recoding site in the genome (Fig. 3). If these recoding sites are G-preferring, we would expect the edited mutants to be normal or even superior to the wild type. Conversely, if the recoding sites prefer A/I, the edited mutants might be defective in tissues or stages where editing occurs, indicating that both versions of the proteins are needed simultaneously. Alternatively, if the edited mutants are normal or even superior in tissues or stages where editing occurs but defective in other tissues or stages where editing does not occur or occurs at lower levels, it suggests that both protein versions need to be expressed in a spatiotemporal manner to resolve trade-offs. While A/I-preferring editing is adaptive, not all G-preferring editing is adaptive. Categorizing G-preferring sites into restorative or diversifying through comparative analysis is crucial, as G-preferring diversifying sites are adaptive (Fig. 1a). However, determining the adaptiveness of G-preferring restorative sites is challenging due to the difficulty in practically differentiating compensatory sites from harm-permitting ones.
If developmental or physiological defects are observed in the edited mutants but not in the uneditable ones, it raises the possibility that editing events might be deleterious (A-preferring). While this suggests that recoding editing has a significant impact on protein function, it does not necessarily reflect its overall importance for the organism, particularly because the unedited version is still expressed in the wild type. Therefore, the availability of only edited mutants is insufficient to determine the developmental or physiological importance of recoding editing.
It is important to recognize that genetic assessments are conservative and may be overly strict. Typically, mutant phenotypes are assessed under standard laboratory conditions, with only a limited set of parameters evaluated. The functional significance of certain recoding sites might be subtle or only become apparent under specific conditions. Biochemical methods that assess the function of unedited and edited versions of proteins in vitro or in cell lines can help determine the impact of recoding editing on protein function and cell fate, offering insights into the mechanisms of recoding editing regulation. However, without genetic evidence, it is challenging to ascertain whether the functional alteration is important for the physiology and development of the organism.
-
A-to-I mRNA editing occurs specifically during the sexual stages in the fruiting bodies (perithecia) of Sordariomycetes, primarily due to the sexual stage-specific expression of the mRNA editing-activating factor Ame1[18]. Deletion of the AME1 gene or blocking the interaction of Ame1 with FgTad3 in F. graminearum results in smaller perithecia with no ascus or ascospore formation[18], indicating that A-to-I mRNA editing is indispensable for sexual reproduction.
Genetic studies using both uneditable and edited mutants have demonstrated that individual A-to-I recoding sites in 18 genes are critical for various stages of sexual development in F. graminearum, including ascogenous hypha formation, ascus and ascospore development, maturation, and ascospore discharge[29,42,44,45] (Table 1). The majority of A-to-I recoding events critical for sexual development in F. graminearum involve premature stop codon (PSC) correction[29]. In Neurospora crassa, two PSC editing sites have been identified as essential for sexual development[8]. Additionally, a PSC editing site is required for spore killing in Fusarium verticillioides[46]. PSC editing represents a harm-permitting, restorative type of editing that enables the fixation of UAG premature stop codons during evolution while correcting them during sexual development. This mechanism ensures that genes with PSCs remain inactive during vegetative growth but become functional during sexual reproduction after editing[29]. Edited mutants of two PSC-containing genes (PSC69 and PSC64) in F. graminearum exhibit increased sensitivity to environmental stresses during vegetative growth compared to wild-type and uneditable mutants[29]. This finding suggests that PSC editing mitigates the survival costs associated with reproduction caused by antagonistic pleiotropy, providing a selective advantage by resolving survival-reproduction trade-offs. These observations provide compelling empirical evidence that harm-permitting editing is an adaptive mechanism. Harm-permitting editing likely occurs in genes that are essential for tissues or developmental stages with high editing activity but would be detrimental during stages with low or no editing. This mechanism helps resolve evolutionary trade-offs by allowing these genes to remain inactive or non-functional when unnecessary, while restoring their function precisely when needed.
Table 1. Functionally important A-to-I recoding sites identified in fungi and bacteria through genetic studies.
Taxon Target protein Recoding site1
(editing level)Uneditable mutant Edited mutant Ref. Fungi F. graminearum PSC58 *323W (53%) Defective in ascogenous hypha formation No obvious defects [29] Fungi F. graminearum PSC69 *318W (16%) Defective in ascogenous hypha formation Sensitive to stress during vegetative growth [29] Fungi F. graminearum PSC27 *494W (87%) Defective in ascus formation, and
ascus and ascospore maturationNo obvious defects [29] Fungi F. graminearum PSC10 *64W (96%) Defective in ascus formation No obvious defects [29] Fungi F. graminearum PSC20 *263W (91%) Defective in ascus formation No obvious defects [29] Fungi F. graminearum PSC24 *172W (80%) Defective in ascus formation No obvious defects [29] Fungi F. graminearum PSC07 *87W (95%) Defective in ascus formation and discharge No obvious defects [29] Fungi F. graminearum Puk1/PSC03 *576W (86%);
*577W (98%)Defective in ascospore formation
and dischargeNo obvious defects [5,29] Fungi F. graminearum Amd1/PSC04 *221W (97%) Defective in ascus maturation No obvious defects [29,45] Fungi F. graminearum PSC37 *816W (85%) Defective in ascospore formation No obvious defects [29] Fungi F. graminearum PSC17 *34W (89%) Defective in ascospore formation No obvious defects [29] Fungi F. graminearum PSC30 *63W (75%) Defective in ascospore formation No obvious defects [29] Fungi F. graminearum PSC52 *693W (73%) Defective in ascospore formation No obvious defects [29] Fungi F. graminearum PSC64 *419W (63%) Defective in ascus formation, and
ascus and ascospore maturationSensitive to stress during vegetative growth [29] Fungi F. graminearum FgAma1/PSC33 *387W (67%) Defective in ascospore morphogenesis No obvious defects [29,44] Fungi F. graminearum Cme5 R986G (71%) Defective in ascus and ascospore formation No obvious defects [42] Fungi F. graminearum Cme11 T304A (50%) Defective in ascospore formation Defective in ascospore formation [42] Fungi F. graminearum Tub1 N347D (41%) No obvious defects Defective in ascospore formation [84] Fungi F. verticillioides SKC1 *71W (84%) Defective in spore killing — [46] Fungi N. crassa Stk-21 *508W (94%) Defective in ascospore maturation
and germination— [8] Fungi N. crassa NCU10184 *220W (88%) Defective in ascospore formation — [8] Bacteria E. coli HokB Y29C (28%−93%) Mild toxicity, similar to wild type Elevated toxicity [9] Bacteria X. oryzae FliC S128P (0−25%) Reduced tolerance to oxidative stress Increased motility and improved tolerance to oxidative stress [48] Bacteria X. oryzae XfeA T408A (21%−78%) Grew at a slower rate than wild type under iron deficiency Enhanced ion uptake activity and grew more rapidly under iron deficiency [49] Bacteria K. pneumoniae BadR Y99C (4%−10%) — Reduced autoinducer-2 activity, decreased cell growth during the stationary phase, and decreased virulence [11] 1 The asterisk (*) marks the premature-stop codon UAG. In addition to PSC editing, two conserved missense editing (CME) sites in the CME5 and CME11 genes have been shown to play important roles in sexual reproduction in F. graminearum[42]. Although no defects were observed in the edited mutant of CME5, the pre-editing residue of Cme5 is evolutionarily conserved across diverse classes of Ascomycota, while the post-editing residue is rarely hardwired into the genome[42]. This suggests that the editing site is A/I-preferring, where having an editable A at this site is evolutionarily advantageous compared to an uneditable A or a genomically encoded G. In contrast, the CME site in CME11 confers a 'heterozygote advantage' during ascospore formation[42]. Both uneditable and edited mutants of CME11 are defective in ascosporogenesis, suggesting that the coexistence of both edited and unedited variants is required for optimal function. Notably, the CME site in CME11 is predicted to be a phosphorylation site[42]. A-to-I recoding at this site may balance hypophosphorylated and hyperphosphorylated states.
-
Although A-to-I mRNA editing sites are rare in bacteria, the remarkably high percentage of non-synonymous edits suggests that these recoding events are likely adaptive[9−11]. To date, four A-to-I recoding sites have been genetically characterized in bacteria, with uneditable mutants available for three of them, providing insights into their functional roles (Table 1). One example is the Y29C editing site on HokB[9], a toxin-encoding gene in Escherichia coli. HokB plays an antagonistic role in inhibiting bacterial growth and conferring antibiotic tolerance[47]. Uneditable mutants of HokB exhibit mild toxicity and growth rates comparable to the wild type, whereas edited mutants show significantly higher toxicity and reduced growth rates[9]. This suggests that the edited version of HokB is more toxic. Although the functional importance of Y29C editing remains uncertain due to the lack of observable defects in uneditable mutants, the evolutionary conservation of this editing site and its increased editing levels during culture growth imply an adaptive role[9]. Specifically, at low cell density, E. coli may produce the low-toxicity version of HokB to promote growth, while at high cell density, the high-toxicity edited version enhances antibiotic resistance. Thus, RNA editing may fine-tune the tradeoff between growth and antibiotic resistance, optimizing bacterial survival strategies under different conditions.
Two additional A-to-I recoding sites have been characterized in Xanthomonas oryzae: the S128P editing on the flagellar filament protein FliC and the T408A editing on the ferric siderophore outer membrane receptor XfeA. These edits are induced under specific environmental stresses, with S128P triggered by oxidative stress (H2O2) and T408A by iron-deficient conditions[48,49]. Functional studies of uneditable and edited mutants reveal the adaptive significance of these recoding events. In the case of FliC, uneditable mutants exhibit lower tolerance to oxidative stress, while edited mutants display increased motility and enhanced resistance to H2O2[48]. This indicates that S128P editing is crucial for improving X. oryzae survival under oxidative stress. Similarly, for XfeA, uneditable mutants grow slower than the wild type under iron-deficient conditions, whereas edited mutants demonstrate enhanced iron uptake activity and faster growth[49]. This suggests that T408A editing is important for X. oryzae adaptation to iron scarcity. Notably, under conditions of sufficient iron or absence of oxidative stress, no significant growth differences are observed between uneditable and edited mutants for both genes[48,49], indicating that these recoding sites are G-preferring. However, the editing levels in fliC and xfeA are stress-induced, suggesting that the two protein variants may provide differential advantages under varying conditions. Additional phenotypic assessments of both uneditable and edited strains are necessary to confirm whether this recoding editing plays a role in resolving trade-offs between stress resistance and normal growth.
Another example is the Y99C recoding site in a transcriptional regulator badR of Klebsiella pneumoniae. This editing site is highly conserved across K. pneumoniae strains, indicating that RNA editing may play an important functional role[11]. However, studies so far have focused exclusively on edited mutants, which exhibit reduced autoinducer-2 activity, lower cellular density during the stationary phase, and decreased virulence[11]. To fully understand the functional significance and potential adaptive advantage of this editing site, further investigation, including comparative studies with uneditable mutants, is required.
-
Mammals possess two active members of the ADAR family: ADAR1 and ADAR2[15]. The primary role of ADAR1 is to edit dsRNA from transposable elements, preventing their detection by the innate immune system[15]. Protein recoding through ADAR1-mediated RNA editing is not essential for normal development and homeostasis[50]. In contrast, ADAR2 is primarily responsible for recoding events in mammals. Mice with a homozygous deletion of ADAR2 die several weeks after birth, but this defect can be rescued by a homozygous A-to-G mutation in the genomic DNA at the Q/R recoding site of the AMPA receptor GluA2[51], indicating that the GluA2 Q/R site is the only critical recoding site for ADAR2. While the function of recoding editing has been experimentally validated at a few sites (Table 2), most transgenic mice with reduced editing show no obvious defects, except at the GluA2 Q/R site. Recent genetic studies in mice suggest that, apart from the Q/R site, all other recoding edits are dispensable for mammalian homeostasis[52]. Conversely, mice with excessive editing at some sites appear normal or exhibit clear deficiencies, supporting the notion that recoding events in mammals are generally non-adaptive.
Table 2. Functions of genetically characterized A-to-I recoding sites in animals.
Taxon Target protein Recoding site
(editing level)Uneditable mutant Edited mutant Ref. Mice GluA2/ GluRB Q586R (~99%) Exhibiting severe dendritic deficits and early death No significant deficiencies in brain development, health, appearance, and lifespan, yet exhibiting increased hippocampal spine density [54,56,57,85] Mice FLNA Q2341R (87.8% + 3.7%) Normal life expectancy with no apparent abnormalities, yet increased vascular contraction leads to elevated blood pressure and cardiac remodeling Cell lines exhibiting increased stiffness and adhesion, yet impaired migration [62,63] Mice 5-HT2CR I156V/M (4%−100%); N158S/D/G (10%−84%); I160V (22%−96%) Developed normally Severe weight loss, increased sympathetic activity, and higher energy expenditure [60,86] Mice GluK2/ GluR6 Q621R (~76%) Normal behavior but increased susceptibility to seizures from kainic acid − [59] Mice CaV1.3 I/M (45%); Y/C (29%) Enhanced learning ability and long-term memory in mice due to increased Ca2+ influx in hippocampal CA1 pyramidal neurons − [61] Mice CAPS1 E1250G (15%−70%) − Enhanced short-term depression at inhibitory synapses and lean phenotype from increased energy expenditure due to physical hyperactivity [64,87] Mice GluK1/GluR5 Q636R (~50%) − Normal development and behavior [65] Drosophila Adar S458G (~50%) Abnormal behavior Abnormal behavior [67] Drosophila GluClα I27V (80%−94%) Impaired olfactory responses and pheromone-dependent interactions, decreased female receptivity, yet intact motor activity and extended lifespan − [69] The Q/R editing on GluA2 is evolutionarily conserved in vertebrates but absent in Drosophila, suggesting it may be a vertebrate innovation[53]. In adult mouse brains, the editing level at the Q/R site on GluA2 is about 99%[54,55]. Mice engineered with an uneditable GluA2, where the ECS was replaced by the neomycin gene (GluA2neo/neo), showed severe dendritic deficits, and died by postnatal day 20[56], highlighting the critical role of Q/R editing. Conversely, mice with a G-to-A mutation at the Q/R site (GluA2R/R) appeared normal, with no brain deficiencies and even increased dendritic spine density compared to wild-type mice[54,57]. These observations, along with nearly complete editing at the Q/R site, support the idea that this editing site has a restorative function. However, recent studies indicate that the edited GluA2R is the ancestral state[53]. The absence of genomic fixation of a potentially advantageous G at this site in vertebrates suggests a preference for A-to-I editing. Attempts to determine the role of unedited GluA2Q in specific regions of the central nervous system have been unsuccessful[58]. Given the predominant expression and editing of GluA2 in the brain[53], we speculate that while GluA2R is essential for the central nervous system, its expression in non-nervous tissues might be detrimental. In line with findings on recoding editing in fungi for resolving survival-reproduction trade-offs[29], Brain-specific Q/R editing may offer a selective advantage over a genomically encoded G by reducing the negative effects of GluA2R expression outside the central nervous system. Further investigation into whether edited mutants exhibit any defects in non-nervous tissues could illuminate the adaptive advantages of the Q/R site on GluA2.
In addition to the Q/R site on GluA2, mice deficient in RNA editing have been engineered for several other proteins, including the glutamate receptor GluK2/GluR6[59], the serotonin receptor 5-HT2CR[60], the calcium channel CaV1.3[61], and the actin crosslinking protein FLNA[62]. Transgenic mice with impaired FLNA Q2341R editing exhibit increased smooth muscle contraction, leading to elevated blood pressure and cardiac remodeling[62]. This highlights the crucial role of Q2341R editing in maintaining cardiovascular health. Further cellular assays reveal that the unedited version (FLNAQ) reduces cellular stiffness and adhesion while enhancing cell migration, whereas the edited version (FLNAR) increases cellular stiffness and adhesion but impairs cell migration[63]. This suggests that Q2341R editing on FLNA likely provides a selective advantage by balancing cell migration with stiffness and adhesion. Mice with defective Q621R editing on GluK2 behave like wild-type mice but are more susceptible to kainic acid-induced seizures, suggesting that Q621R editing may modulate synaptic plasticity and seizure vulnerability[59]. However, whether this editing site is G-preferring or A/I preferring remains unknown. Notably, uneditable mice (CaV1.3ΔECS) lacking the I/M and Y/C editing on CaV1.3 demonstrate improved learning and enhanced long-term memory, aligning with decreased editing levels during learning[61]. This suggests that loss of CaV1.3 RNA editing may enhance hippocampal plasticity, learning, and memory[61]. To understand the importance of RNA editing, it's crucial to identify defects in uneditable mutants. While better performance than wild-type mice might imply that editing events are deleterious, the evolutionary conservation of CaV1.3 RNA editing[61] makes this unlikely. Further investigations into defects in fertility or tissues with up-regulated editing levels could shed light on the adaptive advantages of CaV1.3 RNA editing. Fully edited 5-HT2CR mice exhibited a severe reduction in body fat due to constant activation of the sympathetic nervous system and increased energy expenditure, while mice with blocked 5-HT2CR editing showed no obvious phenotype[60]. This suggests significant impacts of editing on 5-HT2CR function, though its overall importance remains unclear, as the unedited version is present in wild-type mice, raising the possibility that editing could be deleterious.
Only edited mice were available for CAPS1 and GluK1/GluR5. Sole expression of edited CAPS1 caused hyperlocomotion in mice, leading to increased energy expenditure and leanness[64]. Without uneditable mice, it is difficult to determine whether both versions of CAPS1 are necessary or if editing is deleterious. Since edited GluR5 mice do not exhibit developmental or behavioral issues, it has been suggested that GluR5 editing may not be crucial[65]. However, to truly determine the functional importance of these editing sites, it is essential to examine defects in uneditable mutants rather than in edited ones.
-
Recoding editing sites are relatively enriched in Drosophila[20,21]. Currently, the in vivo functions of two genetically characterized recoding editing sites have been identified (Table 2). The first is the S458G editing site in the deaminase domain of Adar, which reduces the catalytic activity of Adar[66,67]. Both fully edited AdarG and uneditable AdarS mutant flies show abnormal behavior[67], indicating that both excessively high (unedited version) and excessively low (edited version) Adar activity is detrimental. RNA editing balances Adar activity by enabling the simultaneous expression of both protein versions. The evolutionary maintenance of the S458G editing site[68] supports its adaptive advantage.
The second recoding site is the I27V editing site in the glutamate-gated chloride channel (GluClα)[69]. This site is evolutionarily conserved and exhibits high levels of editing in neuronal populations. Flies lacking the I27V edit in GluClα show reduced olfactory responses to odors and impaired pheromone-dependent social interactions[69]. Interestingly, while these unedited flies maintain intact motor activity and even have an extended lifespan, GluClα unedited females exhibit significantly longer latencies to copulate, indicating reduced receptivity[69]. I27V editing on GluClα likely provides an adaptive advantage for Drosophila by balancing reproduction and lifespan. Further studies to determine whether edited mutants exhibit enhanced receptivity but reduced lifespan would confirm the adaptive role of the I27V editing in resolving reproduction-lifespan trade-offs.
-
A-to-I recoding, constrained by the genetic code, often results in the substitution of amino acids with smaller side chains, which typically exhibit reduced stability[37,70]. This process primarily occurs in the nervous systems of animals, indicating that the resulting protein destabilization or increased flexibility is crucial for rapid responses to fluctuating external environments. Supporting this, in vitro studies of specific recoding sites in nervous system-related genes suggest that A-to-I recoding fine-tunes protein function by modulating stability, activity, and protein-protein interactions (Table 3). For example, at the Q/R sites of GluA2, GluK1, and GluK2, A-to-I recoding has been shown to reduce calcium ion permeability and channel conductivity[65,71,72]. Specifically, the edited version of GluA2 reduces calcium influx and rescues lethality in ADAR2 knockout mice[51]. Similarly, the I400V recoding in the human potassium channel Kv1.1 decreases side chain size, weakens hydrophobic interactions, and accelerates recovery from the inactive state[73]. A comparable effect has been observed for the I/V recoding of potassium channel Kv2 (Shab) in Drosophila melanogaster[74]. Other recoding events in potassium channels of Drosophila and cephalopods also tend to destabilize specific interactions or states, further supporting this pattern[74−79]. Notably, some of these recoding sites are evolutionarily conserved[51,61,67,80], suggesting that A-to-I recoding may confer adaptive advantages compared to the genomically encoded G. One possibility is that recoding resolves the trade-off between the differing stability requirements of proteins across tissues or conditions. Alternatively, A-to-I recoding may balance excessive protein stability and instability by allowing the simultaneous expression of both edited and unedited variants.
Table 3. Impact of A-to-I recoding on protein function in animals.
Taxon Target protein Recoding site Impact on protein function Ref. Mice GluA2/GluRB Q586R Reduced Ca2+ permeability and channel conductance [71] Mice GluK1/GluR5 Q636R Reduced Ca2+ permeability and channel conductance [65] Mice GluK2/GluR6 Q621R Reduced Ca2+ permeability and channel conductance [72] Mice 5-HT2CR I156V+N158S+I160V Decreased interaction with G proteins and agonist potency [88,89] Mice CaV1.3 I/M+Q/R+Y/C Reduced CaM binding and calcium-dependent inactivation (CDI) [90,91] Rat GluA2/GluRB R764G Enhanced recovery rate from desensitization [92] Rat GluA3 R769G Enhanced recovery rate from desensitization [92] Rat GluA4 R765G Enhanced recovery rate from desensitization [92] Human GABAA (α3) I314M Probably destabilized the stability of the open ion state [80] Human Kv1.1 I400V Destabilized the fast inactivated state and reduced whole-cell
current by decreasing surface membrane trafficking[73,93] Human NEIL1 K242R Reduced activity in removing oxidized pyrimidines [94,95] Human SLC22A3 N72D Reduced direct binding to ACTN4 [96] Human COPA I164V Reduced protein stability and altered conformation [97] Human GLI1 R701G Reduced transcriptional activity and less susceptible to
inhibition by the negative regulator[98] Human AZIN1 S367G Increased protein stability and caused AZIN1 translocation from cytoplasm to nucleus [99] Human RhoQ N136S Enhanced RhoQ activity [100] Human IGFBP7 K95R Increased IGFBP7 protein stability [101] Octopus synaptotagmin-1 I248V Decreased the binding affinity for Ca2+ and altered the protein's conformation [82] Octopus kinesin-1 K282R Reduced transport velocity and run length [82] Octopus Kv1.1 I321V Destabilized the channel's open state [75] Squid Kv1.1 R87G Destabilized the tetramer [76] Squid Na+/K+ ATPase (α1) I877V Destabilized the Na+-bound state [77] Squid Kv2 Y576C; I597V Affected the rate of channel closure and slow inactivation [81] Squid kinesin S75G + Y77C + N117D + K368R + K483R; K67R + Y77C + N117D + K368R + K479R + K483G + E515G Enhanced kinesin motility in cold conditions [83] Drosophila Eag K467R + Y548C + N567D + K699R Decreased activation kinetics and minimal inactivation [79] Drosophila Shaker (Kv1.1) T489A Reduced the inactivation rate of the channel [78] Drosophila Shab (Kv2) I681V Probably destabilized the open state [74] Drosophila Adar S458G Decreased catalytic activity and altered Adar nuclear localization [67] -
In poikilotherms, temperature fluctuations pose significant challenges to the coordination of physiological functions. For behaviorally sophisticated coleoid cephalopods, these challenges are amplified due to their complex nervous systems. Cold-adapted proteins generally exhibit greater flexibility, whereas heat-adapted proteins tend to have a higher proportion of larger and more stable side chains[70]. Since A-to-I recoding often reduces protein stability and increases flexibility (Table 3), the resulting edited proteins are naturally better suited to function in low-temperature environments. One particularly common example in coleoid cephalopods is the transformation of isoleucine (I) into valine (V)[75]. For instance, the I321V conversion in Kv1.1 of octopuses destabilizes the channel's open state, allowing Antarctic octopuses to adapt to cold environments[75]. Similarly, in squid Kv1.1, the substitution of arginine with glycine (R87G) within the tetramerization domain disrupts the channel' s ability to form tetramers[76]. Additional examples include the I597V and Y576C editing sites in the Kv2 K+ channel, which modify channel closure and inactivation rates in squid[81], and the I877V substitution in the Na+/K+ pump, which destabilizes the three Na+ ion-bound state, thereby increasing turnover rates at negative voltages. In synaptotagmin-1 of octopuses, the cold-induced I248V editing site decreases Ca2+ binding affinity, further supporting adaptation to cold environments[82]. These modifications, which enhance protein flexibility, enable cephalopods to rapidly sense and respond to environmental changes. Interestingly, RNA editing activity increases significantly at lower temperatures[82,83], potentially enhancing the sensory and response capabilities of cephalopods' nervous systems in cold environments. Beyond the nervous system, squid also utilize A-to-I recoding to produce kinesin variants with improved motility, which may enhance their movement in cold seawater[83]. Thus, the enhanced sensory and motor abilities conferred by A-to-I recoding likely play a crucial role in enabling cephalopods to adapt to colder conditions. Compared to genomically encoded A or G, A-to-I recoding may be more suitable for acclimation and adaptation to rapid environmental temperature changes, allowing cephalopods to maintain optimal physiological performance.
-
This review highlights the critical roles of individual A-to-I recoding sites across fungi, bacteria, and animals, emphasizing the adaptive significance of RNA recoding in resolving evolutionary trade-offs. However, the functional importance and adaptive advantages of many recoding sites remain poorly understood. To advance the understanding of A-to-I RNA editing, several key areas warrant exploration:
Computational and evolutionary analyses: integrating computational predictions with evolutionary conservation studies can help identify candidate adaptive recoding sites. Comparative genomics across species with varying levels of RNA editing activity may reveal patterns indicative of adaptive evolution.
Development of advanced genetic tools: innovative genome-editing technologies, such as CRISPR-Cas systems with base-editing capabilities, should be utilized to create precise mutations that block or mimic RNA editing at specific sites without altering the encoded amino acid sequence. This approach would enable the generation of uneditable or edited mutants.
Comprehensive phenotypic assessments: mutants should be evaluated under diverse environmental conditions and developmental stages to uncover subtle or context-dependent phenotypes associated with RNA editing. In addition to observing physiological and developmental phenotypes, assessing the fitness of mutants is crucial for understanding the adaptive advantages of RNA editing.
Investigating trade-offs and pleiotropy: Research should focus on how RNA editing resolves trade-offs between conflicting physiological demands, such as growth versus stress resistance or reproduction versus longevity. This will improve our understanding of RNA editing as a mechanism for overcoming antagonistic pleiotropy.
As experimental tools and technologies continue to improve, the field is poised to uncover the full spectrum of functions mediated by this versatile molecular mechanism.
-
Not applicable.
This work was supported by grants from the National Natural Science Foundation of China (Grant Nos 32170200 and 32400168) and the Postdoctoral Research Programs in Shaanxi Province (2023BSHTBZZ25).
-
The authors confirm contribution to the paper as follows: study conception and design: Liu H, Xin K; data collection: Xin K, Kang X, Liu M, Fan J; data interpretation: Liu H, Xin K, Du Y, Feng C; draft manuscript preparation: Liu H, Xin K. All authors reviewed the results and approved the final version of the manuscript.
-
All data generated or analyzed during this study are included in this published article.
-
The authors declare that they have no conflict of interest.
- Copyright: © 2025 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Xin K, Kang X, Liu M, Fan J, Du Y, et al. 2025. Navigating trade-offs: the adaptive significance of A-to-I RNA editing in fungi, bacteria, and animals. Epigenetics Insights 18: e002 doi: 10.48130/epi-0025-0002
Navigating trade-offs: the adaptive significance of A-to-I RNA editing in fungi, bacteria, and animals
- Received: 29 November 2024
- Revised: 09 January 2025
- Accepted: 16 January 2025
- Published online: 18 February 2025
Abstract: A-to-I RNA editing is a widespread epitranscriptomic mechanism that diversifies RNA transcripts by converting adenosine to inosine. This process, catalyzed by specific deaminases, mimics A-to-G mutations at the RNA level, often resulting in protein recoding. It occurs in animals, fungi, and bacteria, with regulation that is often specific to tissue, developmental stage, or environmental conditions. For example, in animal nervous systems, fungal sexual development, and bacterial stress responses, A-to-I editing plays a crucial role. This editing mechanism enhances protein diversity and flexibility, helping organisms mitigate evolutionary trade-offs across varying tissues and conditions. Evolutionary studies suggest that conserved recoding events in animals are likely adaptive, whereas in fungi, recoding events tend to be generally adaptive. Research has shown that A-to-I editing is vital for sexual reproduction in fungi, providing a selective advantage by alleviating survival-reproduction trade-offs. In bacteria, A-to-I editing fine-tunes the balance between growth and stress resistance. While the adaptive role of A-to-I editing in animals remains debated, it is recognized for its significance in physiological processes such as reproduction and lifespan in Drosophila, as well as nervous system function in cephalopods. Overall, A-to-I RNA editing expands the functional repertoire of the genome, facilitating adaptation to diverse environments.
-
Key words:
- A-to-I RNA editing /
- Protein recoding /
- Adaptive evolution /
- Tradeoff /
- Deaminases












