-
Non-coding (nc) RNAs are a significant regulatory component of eukaryotic transcriptomes, and while they have little or no protein-coding potential, they demonstrate functional capabilities[1]. Based on the origin and biogenesis, canonical plant ncRNAs can be categorized into various classes, including long ncRNAs (lncRNAs), and small RNAs (sRNAs) such as small-interfering RNAs (siRNAs), and microRNAs (miRNAs)[2,3]. Among ncRNAs, sRNAs are acknowledged as key molecules for maintaining plant genome integrity orchestrating the regulation of plant development and stress responses. A significant variation observed among different classes of sRNA processing and regulation is their origin from different dsRNA precursors; e.g., miRNA precursors are derived from single-stranded RNA (ssRNA) transcripts encoded by the endogenous non-protein coding miRNA genes (MIRs), folding into an imperfect hairpin-like structure[4]. The biogenesis of miRNA begins with the transcription of miRNA genes through RNA polymerase II (Pol II), resulting in the formation of primary miRNA transcripts via DICER-like 1 (DCL1). Subsequently, DCL1 processes precursor miRNAs into miRNA duplexes within the nucleus. Following transfer from the nucleus to the cytoplasm, miRNA, facilitated by ARGONAUTE (AGO), regulates mRNA cleavage[5,6].
MiRNAs are crucial regulators in a wide range of cellular processes, including plant growth, development, response to biological and environmental stressors, and aging[7−9]. The mode of action and biosynthesis of miRNAs on target genes differ significantly. The regulatory pathways of miRNAs are interconnected and work either synergistically or counteract each other to regulate target gene expression, providing genome stability[10]. The interaction of miRNAs and their targets differ divergently across organisms and exhibit spatial and temporal variations in differential expression. As part of the regulatory network, miRNAs closely interact with target genes, including various transcription factor (TF) genes such as APETALA 2 (AP2), Growth-Regulating Factor (GRF), NAM, ATAF1, and CUC2 (NAC), Nuclear transcription factor Y (NF-Y), MYB, TEOSINTE BRANCHED/CYCLOIDEA/PCF (TCP), SQUAMOSA-promoter binding TF (SPL), and WRKYGQK TF (WRKY) etc.[11]. The interplay between miRNA and regulatory components holds significance in governing plant developmental processes, playing a vital role in the modulation of gene expression[12].
The Poaceae family, encompassing grasses, stands as one of the largest families of flowering plants, exhibiting adaptability to diverse climatic zones of all continents[13]. Grasses, as pioneering plants, inhabit disturbed areas that may be unsuitable for other plants to grow. The unique position of grasses among flowering plants is determined by their significant economic contributions[14,15].
Beyond their role in food production, grasses serve as vital sources for constructing materials, contributing to paper and biofuel production. Wild grasses, integral to the vegetation of steppes, pastures, and natural meadows, hold significance as fodder plants. The widespread distribution of grasses suggests their high adaptation to adverse environmental conditions, suggesting the presence of efficient resistance mechanisms. Like other plant species, grasses also rely on miRNAs to regulate important developmental processes. Despite substantial genomic resources, experimental data, and an increasing array of computational tools, the characteristics and functions of miRNAs in grasses remain inadequately understood, indicating a knowledge gap in this domain. In this review, we delve to establish contemporary insights into the biogenesis and biological significance of miRNAs in grasses. We explore the regulatory hierarchy of miRNAs in grasses growth and developmental responses, particularly regarding endogenous and exogenous cues in shaping the functional diversification of gene-regulatory miRNA pathways. We also discuss recent advances in generating miRNAs and their possible biological benefits to plants, specifically grasses.
-
Endogenous miRNA genes (MIRs) encode miRNAs, crucial gene expression regulators at post-transcriptional levels, regulating plant development[4]. Mainly, MIRs are located in the intergenic, non-coding loci in plants, where only a minor portion is located at the subset in the intronic region of the protein-coding genes. Most miRNAs are transcribed from DNA sequences by DNA-dependent RNA polymerase II (Pol II) into single-stranded, polyadenylated, self-complementary RNA molecules, primary miRNAs (pri-miRNAs), which can fold into hairpin-like structures. The self-complementary and imperfect stem-loop pri-miRNAs are processed and cleaved by DCL1 to produce mature miRNA duplexes. The pre-miRNAs undergo processing by DCL1 to generate the imperfect miRNA/miRNA* duplexes 2nt 3' overhangs at each end in the nucleus[16].
Consequently, one strand of the miRNA/miRNA* duplex is selected as the guide strand, and in most cases, is integrated into ARGONAUTE-1 (AGO1), forming the miRNA-induced silencing complex (miRISC). Meanwhile, the other strand, referred to as the passenger strand (miRNA*), is eliminated (Fig. 1). It is worth noting that plant miRNAs undergo a crucial modification for their stabilization and protection from degradation. This modification is known as 2-O-methylation facilitated by the methyltransferase protein HUA ENHANCER 1 (HEN1)[17]. Once methylated, these miRNAs are then transported to the cytoplasm by Hasty (HST) protein for further processing and regulation of gene expression[18]. In the cytoplasm, miRNA* is often considered as non-functional and, therefore, degraded. Simultaneously, the mature miRNA strand, prevailing in abundance, integrates into an RNA-induced silencing complex (RISC) comprising various proteins, notably the catalytic AGO. RISC primarily modulates target gene expression by either inhibiting translation or by cleaving complementary target mRNAs[19]. Translation inhibition occurs when there is low complementarity between the miRNA and its target mRNA, whereas mRNA cleavage requires high complementarity between miRNA and its target mRNA[20]. Nevertheless, miRNAs have been discovered alternatively regulating targets via DNA methylation in rice and additional grasses, indicating versatile manners of miRNAs to target modulations in plants[21].
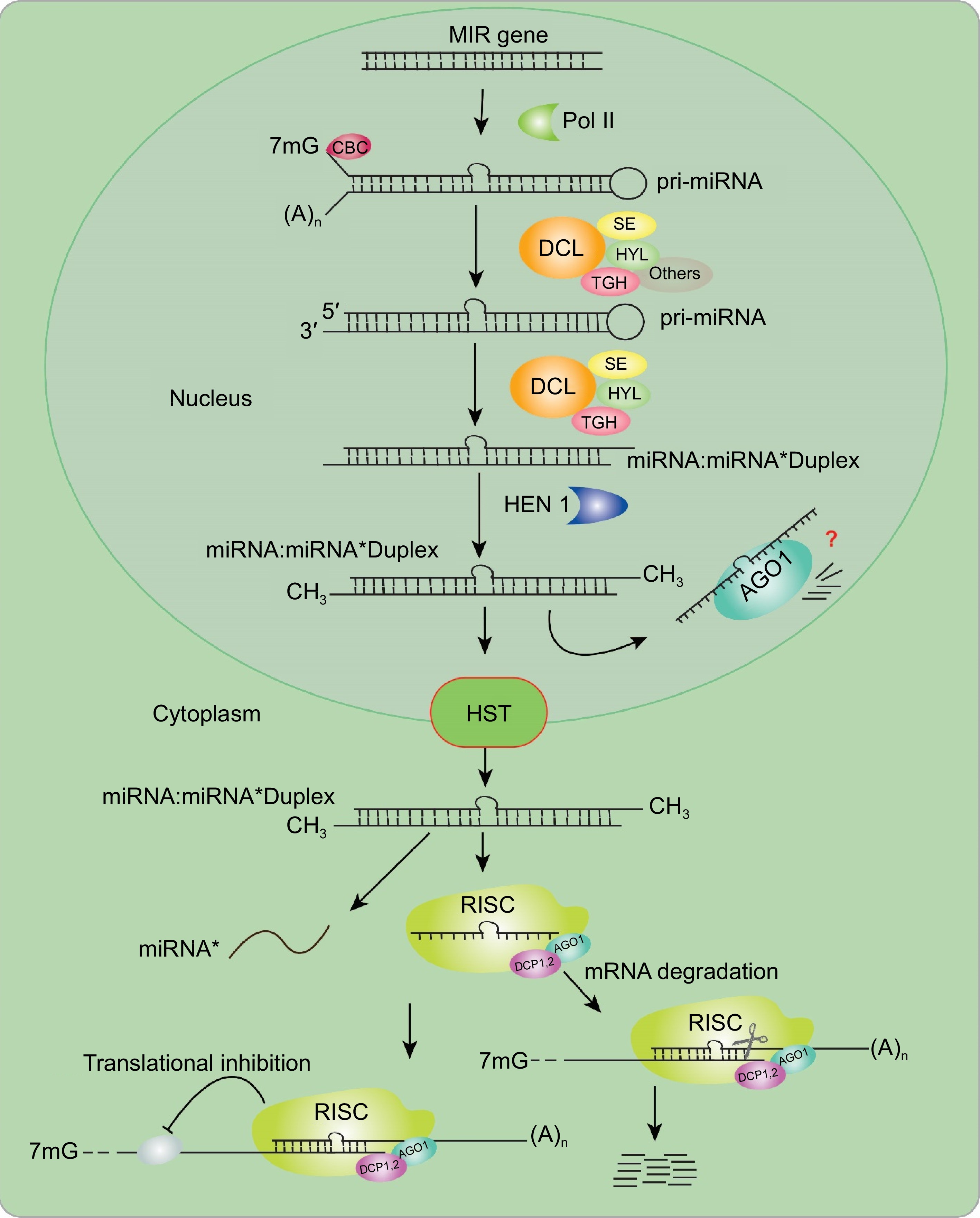
Figure 1.
Biogenesis of model miRNAs in plants. MIR genes are initially transcribed by RNA Pol II into pri-miRNAs, which then adopt hairpin structures through self-folding. The subsequent processing and splicing of miRNAs require the interaction of DDL, HYL1, SE, and TGH in conjunction with the cap-binding complex (CBC). DCL1 processes both pri-miRNAs and pre-miRNAs, generating one or several miRNA/miRNA* duplexes, subsequently methylated by HEN1. These processed miRNAs are transported to the cytoplasm by HST1. Subsequent processing involves the integration of the selected miRNAs into the RISC, which comprises AGO1. This complex guide either the inhibition of translation or the cleavage of the target mRNA transcript. AGO, Argonaute; DCL, Dicer-like; HYL1, HYPONASTIC LEAVES1; Pol II, RNA polymerase II; HEN1, HUA ENHANCER 1; CBC, cap-binding complex; MIR, MIRNA; RISC, RNA-induced silencing complex; miRNA, microRNA; mRNA, messenger RNA; SE, SERRATE.
-
MiRNAs play significant roles in regulating developmental processes in grasses by targeting a variety of genes and TFs involved in distinct regulatory processes, e.g., regulation of meristem properties, leaf development, leaf polarity, flower development, and flowering patterns etc.[22,23]. Variations in the expression of miRNA transcription or processing complexes sometimes have multiple effects on plant growth, form, function, and development, demonstrating the significant importance of miRNAs in plant development. A single miRNA or a miRNA family frequently targets various members of a gene family, showcasing evolutionary conservation in their target genes across related plant species. For example, the miR156-SPL module influences tillering, ear development, panicle branching, and grain size controlling grain production in switchgrass (Panicum virgatum) (Fig. 2)[24,25], depicting the multifaceted regulatory roles of miR156-SPL in initiating axillary meristems in monocots. In barley, the miR171-SCL module is required for the activation of the miR156-SPL pathway during the vegetative phase transition, highlighting a monocot-specific function of miR171-SCL[26]. The coordinated interplay of the two microRNA families, MiR156/157 and miR172, regulates the transition from the juvenile to adult developmental phases and the flowering transition in petunia[27].
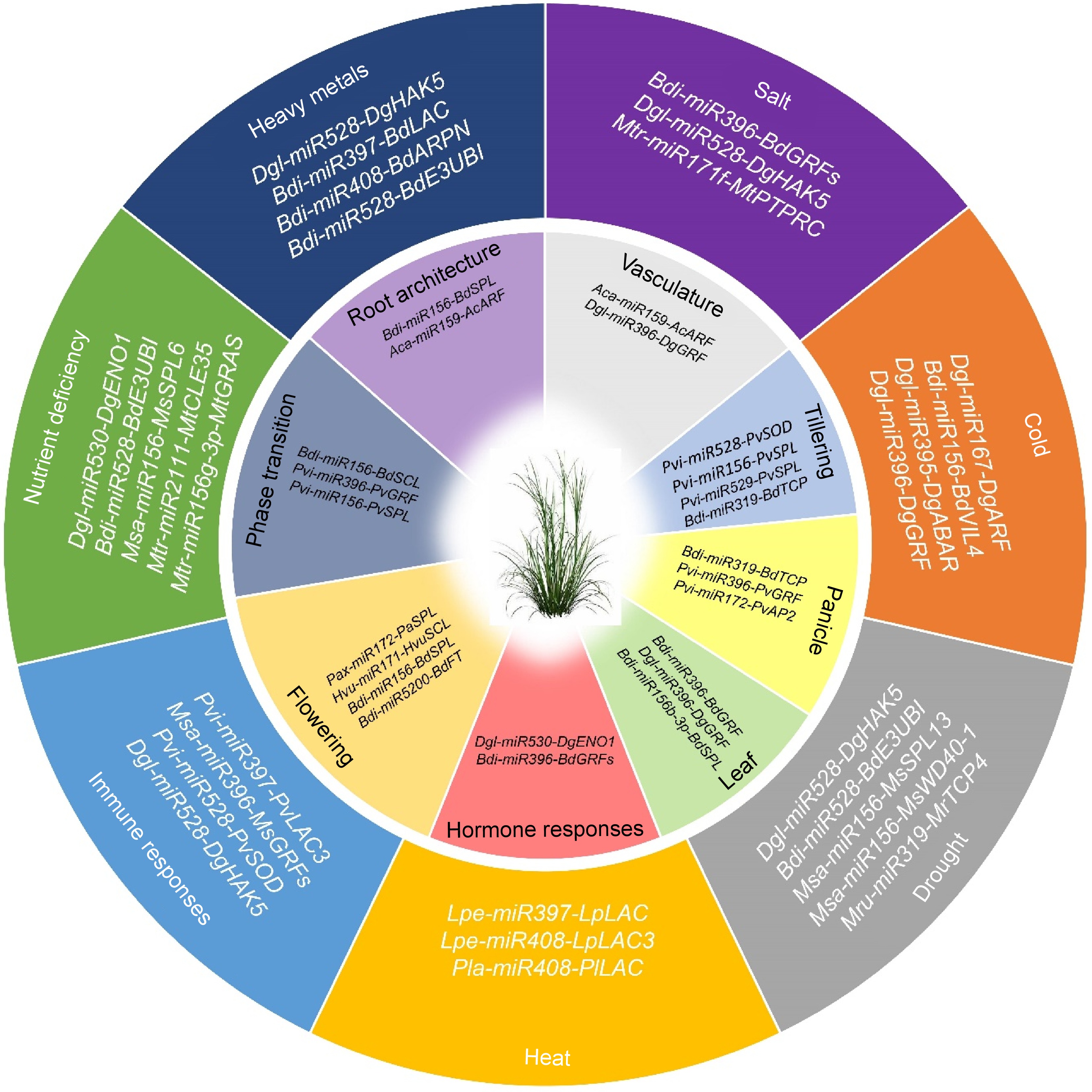
Figure 2.
MicroRNAs play pivotal role in development and stress responses in grasses. Recent understanding of miRNA-mediated regulation is outlined, encompassing development (inner circle) and responses to biotic and abiotic stresses (outer circle) in grasses. Bdi, Brachypodium distachyon; Msa, Medicago sativa; Pvi, Panicum virgatum; Pla, Paeonia lactiflora; Agr, Apium graveolens; Lpe, Lolium perenne; Hvu, Hordeum vulgare; Aca, Agrostis canina; Cda, Cynodon dactylon; Dgl, Dactylis glomerata; Mru, Medicago ruthenica, and Mtr, Medicago truncatula.
Consequently, in most grasses the number of branches determine grain yield. It was observed that the regulation of tillering and panicle branching in switchgrass involved the interplay of miR156/miR529/SPL and miR172/AP2 modules within respective genes sets[24,25]. The SPL gene exerts a negative influence on tillering while concurrently exerting a positive effect on the transformation of the inflorescence meristem and spikelet. Apart from these conserved miRNAs functions, these short non-coding RNA molecules are also involved in the regulation of endogenous and exogenous cues in grasses. In the context of endogenous cues, miRNAs are involved in the fine-tuning of certain developmental processes, influencing growth, differentiation, and reproductive processes by targeting specific mRNA transcripts in grasses. In addition to their involvement in endogenous processes, miRNAs are also responsive to a myriad of exogenous cues, reflecting the adaptability of grasses to changing environmental conditions.
-
While referring to endogenous cues, miRNAs contribute to the fine-tuning of developmental processes ensuring the precise spatiotemporal control of gene expression within grasses. A study investigated the regulatory roles of miRNAs as emerging and active switches in DNA damage response (DDR) and associated pathways in the model forage Medicago truncatula. Various specific miRNAs, such as Mtr-miR156a and Mtr-mir172c-5p, have been investigated for their putative targeting of genes associated with DDR. For instance, Mtr-miR156a is hypothesized to target MtUBE2A, a ubiquitin-conjugating enzyme, involved in histone modification, while Mtr-mir172c-5p is implicated in targeting MtRAD54-like, a protein involved in the repair of double-stranded breaks in DNA. Additionally, Mtr-mir395e is postulated to target DNA methyltransferase 1-associated protein (MtDMAP1), related with histone modifications. Likewise, for DNA-dependent DNA replication, a miRNA, Mtr-miR5741a, is suggested to putatively target E2F TF-E2FE-like protein (MtE2FE-like) involved in DNA-dependent DNA replication[28−31]. Thus, these findings contribute to our understanding of miRNAs in the post-transcriptional regulation of DDR in forages.
Additionally, the conserved mature sequences of several miRNAs in grasses imply their similar roles in regulating target gene activity. For example, the miR528 shares an identical mature sequence in many grass species, such as Brachypodium distachyon, maize, rice, sugarcane, and Sorghum bicolor. The sequence similarity of miR528 across different monocot species implies that this miRNA may function similarly in regulating the activity of target genes[32]. Likewise, another study reported that the inhibition of miR528 by short tandem target mimic (STTM) and quadruple pvmir528 in switchgrass mutants derived from genome editing has resulted in an elevated tiller number and enhanced regrowth following mowing. Degradome analysis and in situ hybridization assay showed that the upregulation of two miR528 targets encoding Cu/Zn-SOD enzymes was responsible for improved regrowth and tillering in pvmir528 mutants[33], illuminating a novel regulatory function of miR528 in controlling biomass traits and contributing to improvements in crop yield.
Although not a biofuel crop, B. distachyon, an extensively studied model grass species, shares similarities with various biofuel feedstocks. Analysis of differentially expressed miRNAs and their respective target genes in B. distachyon contribute to comprehending the mechanisms governing oxidative stress responses and tolerance in grasses[34]. Another study examined the function of miR5200, a B. distachyon miRNA that targets the FLOWERING LOCUS T (FT) orthologs, FT1 and FT2. MiR5200 specifically influences the response to photoperiod with stable levels during vernalization but high levels in SD conditions and indiscernibly low levels in LD conditions. Regulation of miR5200 includes increased H3K27me3 histone modification in LD conditions. Additionally, plants overexpressing miR5200 flower later in LD conditions, while miR5200 target mimics, expressing an uncleavable mRNA complementary to the miRNA, flower earlier in SD conditions. Thus, miR5200 specifically represses FT1 and FT2 in SD conditions, illustrating the regulatory roles of sRNAs to rewire gene networks preventing flowering[35]. Moreover, another study hypothesized that the maturation process of pri-miR5200 would occur uniformly before flowering transition, implying a negative correlation of pri-miR5200 with FT levels. However, if the pri-miRNA processing occurs predominantly in the late vegetative stage, possibly after the juvenile-to-adult transition, and there is co-expression of the pri-miR5200 and FT, it would anticipate a positive correlation between the two transcripts during early development[36], suggesting a developmental contingency explaining the observed positive correlation in Bd21.
Moreover, the model of miRNA and GROWTH REGULATING FACTOR genes (GRFs), miR396-PvGRF is reported to regulate plant height and biomass yield in switchgrass[37]. MiR396 targeted PvGRF1, 3, and 9 showed a higher significant expression in stem (Table 1). The effective restoration of the impaired phenotype in OE-miR396 plants by overexpressing PvGRF9 further underscore the potential of PvGRF9 as a promising molecular tool for enhancing plant biomass yield and feedstock quality[37]. The Corngrass1 (Cg1) gene in maize encodes a miRNA that induces morphology and juvenile cell wall identities. To elucidate its function, the Cg1 gene was transferred and overexpressed in switchgrass. Overexpression of Cg1 miRNA inhibits flowering by increasing starch content in switchgrass[38]. Plants overexpressing Cg1 resulted in up to 250% surge in starch content, consequently enhancing glucose release in saccharification assays, irrespective of biomass pretreatment. This underscores the potential applicability of this approach not only in the domestication of new biofuel crops but also in addressing concerns related to the spread of transgenes into native plant species. However, there is also evidence that the response of miRNAs to the same abiotic stress can also occur in a genotype/species-dependent manner. For example, the expression of miR156 regulates floral transition in B. distachyon under lower temperatures. However, elevated miR156 levels delay reproductive growth in cold periods, while excessive miR156 accumulation in late autumn hinders timely reproduction (Fig. 2)[34]. It is also unknown whether miRNAs can regulate age-dependent events and growth transitions. Therefore, a recent study revealed global alternative splicing variations in miR156-overexpressed B. distachyon plants delayed growth transitions. This study also reported that miR156 targeted SPLs and regulated their expression in intron retention alteration in addition to mRNA clearance and translation inhibition manners[34], illustrating a substantial machinery of alternative splicing in miRNAs that mediates growth transitions in grasses.
Table 1. List of miRNAs involved in grasses development.
miRNA Species Target mRNA Biological function Ref. Bdi-miR156 Brachypodium distachyon BdSPL Root development/Leaf development/Flowering [25] Aca-miR159 Agrostis canina AcARF Root development/Vasculature [52] Dgi-miR396 Dactylis glomerata DgGRF Vasculature/Leaf development [63] Pvi-miR528 Panicum virgatum PvSOD Tillering [33] Pvi-miR156 Panicum virgatum PvSPL Tillering [24,25] Pvi-miR529 Panicum virgatum PvSPL Tillering [24] Bdi-miR319 Brachypodium distachyon BdTCP Tillering/Panicle [61] Pvi-miR396 Panicum virgatum PvGRF Panicle [37] Pvi-miR172 Panicum virgatum PvAP2 Panicle [24,25,27] Bdi-miR396 Brachypodium distachyon BdGRF Leaf development/Hormone response [37] Bdi-miR5200 Brachypodium distachyon BdFT Flowering [35] Hvu-miR171 Hordeum vulgare HvSCL Flowering [26] Pax-miR172 petunia PaSPL Flowering [27] Hence, the coordination of gene expression programs at specific developmental stages by lncRNA-miRNA-targets is plausible. The comprehensive genome-wide characterization of B. distachyon lncRNAs facilitates the rapid identification and confirmation of RNA-based regulatory elements in grasses, serving as potential targets for biotechnological applications. Consequently, a study was undertaken to assess the genetic association between MIRs and lncRNAs in B. distachyon. Interestingly, conserved miR167d and miR399a were discerned in the non-coding regions of the lncRNAs TCONS_00007390 and TCONS_00016338, respectively, implying a spliced intron the origin for these noncoding transcripts. Furthermore, MiR395c and MiR395j were identified within the mono-exonic lncRNA TCONS_00072591, signifying the simultaneous transcription of a polycistronic transcript. Several lncRNAs target MiR156, a key regulator of flowering and leaf development in plants, particularly, the 3′ end of bdi-miR156g, was identified as a potential target of the lncRNA TCONS_00042468 which is highly expressed in Bd21 early inflorescence and pistil. Similarly, TCONS_00067507 exhibits a comparable expression pattern, demonstrating sequence complementarity with bdi-miR156b-3p[39]. Likewise, the lineage-specific miR5174f displays significant complementarity in the target mimic region with multiple lncRNA transcripts, potentially acting to sequester mature miRNA[39].
Some miRNA pathways are also reported to regulate shoot apical meristem (SAM) in grasses. The STM-WUS-CLV pathway regulates cell identity in the SAM and, to a somewhat lesser extent, in the flower meristem. Various miRNAs have been shown to play roles in the synthesis and maintenance of the SAM by affecting targets within the STM-WUS-CLV pathway. The expression of WUSCHEL (WUS) in the organizing center (OC) of the SAM is subject to regulation by miR394, synthesized in the L1 layer, a single-cell layer on the surface of SAM in rice[40]. Altogether, these results illustrated that miRNAs are integral parts of the plant regulatory machinery, orchestrating diverse biological processes for the development and adaptability of plants to cope with environmental challenges.
-
MicroRNAs are complex regulators of gene expression crucial for development, stress responses, and environmental adaptations in grasses. Therefore, to understand the regulatory mechanisms governing miRNA expression, it is pivotal to unravel the intricate roles of exogenous cues that shape plant responses. Here we will explore the world of miRNA regulation in grasses influenced by the surrounding environment.
Resistance genes, also known as R genes, are genes that provide disease resistance against pathogens by producing R proteins. Tight regulation of R genes is required for biotic stress resistance, as their activation in the absence of infection would be deleterious to plants. Therefore, a study in B. distachyon putatively predicted 33 new miRNA sequences, with 9 miRNA families verified through sRNA sequence identification, particularly in disease response genes. Following this, a subset of these miRNAs was evaluated for potential interaction in B. distachyon lines subjected to Fusarium culmorum infection, revealing significant polymorphism in the NBS-LRR family gene, Bradi1g34430, leading to the formation of two major genotypes. One of these genotypes exhibited deletion of its miRNA target sides, thereby inducing altered gene expression during infection[41], suggesting a role of miRNAs in the disease response in B. distachyon. Another study emphasized the crucial role of certain RNAi genes in fungal virulence revealing how miRNAs influence the interaction between B. distachyon and Magnaporthe oryzae (Mo) by deploying miRNA sequencing. This study found various Mo-miRNAs in infected tissues, with a few candidates targeting B. distachyon mRNAs[42], aiming as potential mediators against fungal virulence, further corroborating the critical role of miRNAs in establishing the virulence interaction and its outcome.
Similarly, it was also reported that miR396 was significantly up-regulated in wounding treatment, simulating defoliator insect feeding injury. The MIM396 transgenic Medicago sativa plants exhibited enhanced resistance to Spodoptera litura larvae with elevated lignin contentment but decreased JA accumulation[43]. Moreover, most of the miR396 putative target GRFs were up-regulated in the transgenes and responded to the wounding treatment by providing genetic resources for studying the roles of miRNAs in insect resistance (Table 2). MiRNAs also play a crucial role in maintaining nutrient homeostasis in grasses by orchestrating the expression of transporter genes involved in nutrient uptake and utilization. In this context, a study overexpressed miR156 in M. sativa to check the nutritional profile, energy values, in vitro degradation, and fermentation against SPL6RNAi, SPL13RNAi, and WT. The overexpression of miR156, miR156-OE, exhibited lower fiber content but increased energy values. Chemical analysis revealed that SPL6RNAi, which is more similar to miR156-OE, plays a significant role in the nutritional profiling of M. sativa in the miR156-OE event[44].
Table 2. List of miRNAs involved in responses to biotic and abiotic stresses.
miRNA Species Target mRNA Biological function Ref. Bdi-miR396 Brachypodium distachyon BdGRF Salt [37] Dgl-miR528 Dactylis glomerata DgHAK5 Salt/Drought/Immune response/Heavy metals [33] Mtr-miR171f Medicago truncatula MtPTPRC Salt [52] Dgl-miR167 Dactylis glomerata DgARF Cold [63] Bdi-miR156 Brachypodium distachyon BdVIL4 Cold [62] Dgl-miR395 Dactylis glomerata DgABAR Cold [58] Dgl-miR396 Dactylis glomerata DgGRF Cold [37] Bdi-miR528 Brachypodium distachyon BdE3UBI Drought/Nutrient deficiency/Heavy metals [56] Msa-miR156 Medicago sativa MsSPL13 Drought [60] Msa-miR156 Medicago sativa MsWD40-1 Drought [60] Mru-miR319 Medicago ruthenica MrTCP4 Drought [61] Lpe-miR397 Lolium perenne LpLAC Heat [51] Lpe-miR408 Lolium perenne LpLAC3 Heat [50] Pla-miR408 Paeonia lactiflora PlLAC Heat [47] Pvi-miR397 Panicum virgatum PvLAC Immune response [51] Msa-miR396 Medicago sativa MsGRF Immune response [43] Pvi-miR528 Panicum virgatum PvSOD Immune response [33] Dgl-miR530 Dactylis glomerata DgENO1 Nutrient deficiency [63] Msa-miR156 Medicago sativa MsSPL Nutrient deficiency [44] Mtr-miR2111 Medicago truncatula MtCLE35 Nutrient deficiency [14] Mtr-miR156g-3p Medicago truncatula MtGRAS Nutrient deficiency [45] Bdi-miR397 Brachypodium distachyon BdLAC Heavy metals [51] Bdi-miR408 Brachypodium distachyon BdARPN Heavy metals [51,59] In a study, MtCLE35, a CLAVATA3-like (CLE) signaling peptide, was found up-regulated by high-nitrogen (N) environments, resulting in the inhibition of nodule formation by impacting rhizobial infection. A high-N condition or MtCLE35 expression suppresses the accumulation of miR2111, hindering its positive effects on nodulation. Contrastingly, the downregulation of MtCLE35 by RNAi or the ectopic expression of miR2111 leads to an increased miR2111 accumulation independent of the N environment, and thus partially bypasses the nodulation inhibitory process[14], demonstrating that the N-induced MtCLE35 signaling peptide acts through other receptors and the miR2111 systemic effector to inhibit nodulation.
MiRNAs are also known to play a role in abiotic stress tolerance under adverse environmental conditions by regulating various stress-responsive genes, TFs, and phytohormones. Foxtail millet, genetically closely related to grasses and other bioenergy-related crops, exhibits potential for abiotic stress tolerance. Through a comprehensive genome-wide in silico analysis, 355 mature miRNAs, their secondary structures, and corresponding targets were identified[45]. Predicted targets of these miRNAs were encoding DNA binding proteins, TFs, and important functional enzymes crucial for plant abiotic stress responses that would assist miRNA studies in grass research. Similarly, 55 miRNAs and their respective targets were detected under aluminum stress in Medicago truncatula. Upon further analysis, it was observed that mtr-miR156g-3p significantly upregulated in all treatments directly involved in the expression related to the root cell growth. Additionally, three miRNAs, novel_miR_135, novel_miR_36, and novel_miR_182, collectively regulated the expression of four aluminum-related TFs, including GRAS, MYB, WRKY, and bHLH, implying their involvement in responses to aluminum stress[45]. Furthermore, miR2119 and miR5213 were implicated in aluminum stress tolerance by modulating F-box proteins, known for their protective roles against stress, further contributing to a deeper comprehension of the role of miRNAs in grasses response to aluminum stress[46].
Recent observations revealed that heat induces the expression of miR408 in herbaceous peony (Paeonia lactiflora), with notably higher levels in heat-resistant cultivars, suggesting a potential role for miR408 in the development of heat stress tolerance[47]. Similarly, miR408 expression in celery (Apium graveolens) was increased upon exposure to high temperatures[48]. Contrastingly, a study in switchgrass reported that heat stress led to a downregulation of miR408 expression[49]. Despite disparate findings regarding the response of miR408 to high temperatures in various grass species, there is a lack of experimental evidence elucidating its function in plant heat stress[50]. Transgenic perennial ryegrass (Lolium perenne) overexpressing rice miR408 exhibited heightened heat stress tolerance through the suppression of laccase (LAC3) gene expression. The elucidation of the molecular mechanism through which miR408 enhances plant tolerance to high temperatures has the potential to yield valuable insights, introducing novel concepts and genetic resources essential for breeding heat-resistant cool-season turfgrass in future research[50]. During heat stress, MeJA-primed plants exhibited miR397 and novel-m0008-5p-regulated expression of laccase genes (LpLAC and LpLLI) in switchgrass. MeJA likely enhances heat tolerance by inducing miR397 and novel-m0008-5p expression, thereby impeding lignin degradation through laccase inhibition[51].
In a plant species closely related to bermudagrass, miR319 overexpression in creeping bentgrass led to a significant improvement in salt tolerance. This enhancement was attributed to an increase in photosynthetic performance. Interestingly, a few miRNAs, miR171f, miR319, miR156, and miR159 have conserved targets in the photosynthesis-related pathway by specifically targeting the NAD+ binding and electron transport photosynthesis. Overall, these findings imply that the miRNAs may contribute to the regulation of photosynthesis by transcriptional repression under salt stress[52]. A study in switchgrass identified conserved miRNA families, miR156, miR442, miR1171, miR1423, miR1869, miR2199, miR3436, and miR5140 which comprised the top largest miRNAs in switchgrass. Certain miRNA families, such as miR156, miR442, miR1171, and miR2199, exhibited stable expression across three libraries, CK, salinity, and drought. In contrast, miR1869 counts diverged significantly in the CK, drought, and salinity libraries, suggesting distinct roles and responses of miRNAs in grass development under different conditions[53]. High abundance in members of conserved miRNA families, such as miR156, miR172, miR166, miR168, miR167, and miR444, indicates that the overexpression of these miRNAs could potentially contribute to the maintenance of normal biological functions in bermudagrass (Cynodon dactylon) libraries[52].
In the same manner, transgenic creeping bentgrass overexpressed Osa-miR396c to elucidate its role in perennial grass. Transgenic creeping bentgrass targeted by miR396 displayed modified development characterized by reduced plant height and reduced number of epidermis cells per unit area compared to WT. Moreover, these transgenic plants exhibited enhanced salt tolerance, linked to improved water retention, thereby providing insights into the developmental regulatory networks mediated by miRNAs[54]. Another study utilized four sRNA libraries from the roots of Medicago truncatula and Medicago sativa and investigated their regulation under salt stress by discovering 385 conserved and 68 new candidate miRNAs from 96 families. miRNAs identified under salt stress had a wide range of targets some of which displayed contrasting expression patterns between M. truncatula and M. sativa roots revealed by statistical analysis of the abundance of sequencing reads, which might play important roles in salt stress regulation in Medicago[55]. Similarly, the overexpression of miRNA171f, targeting pentatricopeptide repeat-containing protein in M. truncatula, led to an increased photosynthetic performance through transcriptional repression of genes involved in the electron transport pathway under salt stress, suggesting a potential target for future breeding in grasses[52].
A multifaceted miRNA, miR528, emerges as a pivotal miRNA in grasses responding to water and oxidative stress. Consequently, drought stress elevated the expression of bdi-miR528 in Bd21, underscoring the potential significance of miR528[56]. In B. distachyon, bdi-miR528 targets Bradi3g19170, a gene encoding E3 ubiquitin-protein ligase, is potentially involved in ubiquitination and ethylene signaling pathways during abiotic stress responses[57]. Another study in B. distachyon reported 39 known and 221 novel miRNAs, among which 31 known miRNAs and 30 novel miRNAs were related to the response to and resistance against H2O2-stress in Bd21. The complex interplay of these miRNAs offered valuable insights into the mechanism of signal transduction and oxidative stress resistance mechanisms under H2O2 stress[58]. Furthermore, the MIR395 gene family represents the largest MIR family and includes 15 members, in which 13 members were identified to be overexpressed under H2O2 stress, with their target mRNAs were associated with oxidative stress responses[58]. Similarly, the identified members of the MIR395 gene family exhibited upregulation in B. distachyon during drought stress[56], further verified the multifaceted roles of miRNAs in grasses development.
In perennial ryegrass overexpressing miR408 exhibited accelerated growth under drought stress compared to the control, with more pronounced stress signals. The miR408-OE induced leaf morphological changes that contributed to reduced water loss and heightened antioxidative capability in the ryegrass, revealing specific and drought-responsive characteristics of miR408. Therefore, miR408 is considered a potential target for genetic modifications aimed at enhancing water stress tolerance in perennial grasses. Hence, these miRNAs exemplify the regulatory potentials of miRNAs in modulating crucial plant processes, generating novel short regulatory RNAs, and restructuring gene regulation networks[51,59], further investigations across diverse perennial grass species would be advantageous for comprehensive understanding and application.
The interplay between miR156-regulated SPL13, and DIHYDROFLVONOL-4-REDUCTASE (DFR) regulating WD40-1 could be another mechanism of miRNAs to regulate drought stress by the relative expression of genes related to metabolite and physiological strategies. A study reveals that moderate to low relative miR156 transcript levels are required for enhanced drought resistance in M. sativa by silencing SPL13 and enhancing WD40-1 expression, while higher miR156 overexpression resulted in drought susceptibility[60]. Likewise, miR319 was found to significantly downregulate the expression of teosinte branched/cycloidea/proliferating cell factor 4 (TCP4), encoding plant-specific transcription factors, significantly in the leaves, further illustrating that the miR319-TCP module can act as a homeostasis mechanism in Medicago ruthenica roots following drought stress and is conserved among plant species[61].
A VERNALIZATION INSENSITIVE 3 (VIN3)-like gene, BdVIL4, in B. distachyon, preferentially modulates miRNA156 spatially expressed in young tissues. Plants with reduced BdVIL4 exhibit phenotypes similar to miRNA156 over-expressors[62], indicating that BdVIL4 prevents excessive response to cold temperatures by suppressing the accumulation of miR156. The intricate regulatory roles of miRNAs in orchardgrass (Dactylis glomerata) extend through the vernalization and heading stages. In this regard, a study identified 69 differentially expressed miRNAs across five flowering stages, revealing significant variations in the expression of miR395, miR530, miR167, miR396, miR528, novel_42, novel_72, novel_107, and novel_123 during vernalization[63]. The targets of these miRNAs were associated with transmembrane transport, phytohormones, and plant morphogenesis during vernalization, providing valuable insights for genetic engineering to regulate flowering in orchardgrass (Fig. 2). Altogether, the regulatory influence of miRNAs in grasses is beyond the endogenous and exogenous domains underscoring the versatility of miRNAs as molecular switches to balance internal developmental processes with external environmental cues for optimal survival and adaptation of grasses.
-
In the realm of miRNAs, extensive research has explored the conservation of sRNAs across plant species, including grasses, revealing highly conserved miRNA families and their targets, indicative of shared functional regulatory networks. Nonetheless, a significant proportion of miRNA sequences are species-specific, indicating the existence of numerous young or emerging miRNAs. In contrast, our understanding of miRNA conservation in grasses is relatively limited, highlighting the complexity and diversity of miRNA biology across grass taxa. Emerging evidence has demonstrated the crucial regulatory roles of miRNAs in grass development, involving DCLs, RDRs, and AGO proteins in miRNA pathways. However, the downstream modulation of these regulators concerning plant development remains largely unknown. Various questions raised in this study include: (i) How do miRNAs regulate reproduction in grasses? (ii) How do miRNAs bridge the gap between endogenous developmental stimuli to external environmental changes? (iii) Have the roles of miRNAs in grasses evolved to meet specific ecological challenges? To answer these questions, researchers must exert more effort and resources to explore the developmental roles of miRNAs in grasses.
-
The authors confirm contribution to the paper as follows: conceptualization, visualization, writing – original draft, review and editing: Sajid M; conceptualization, software: Fahad M; investigation, supervision: Zhou W; conceptualization, supervision, review and editing: Wu L. All authors reviewed the results and approved the final version of the manuscript.
-
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
This work was supported by the Sanya Science and Technology Innovation Program (2022KJCX48).
-
The authors declare that they have no conflict of interest. Liang Wu is the Editorial Board member of Grass Research who was blinded from reviewing or making decisions on the manuscript. The article was subject to the journal's standard procedures, with peer-review handled independently of this Editorial Board member and the research groups.
- Copyright: © 2024 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Sajid M, Fahad M, Zhou W, Wu L. 2024. MicroRNA regulation and environmental sensing in grasses. Grass Research 4: e012 doi: 10.48130/grares-0024-0010
MicroRNA regulation and environmental sensing in grasses
- Received: 14 January 2024
- Revised: 02 March 2024
- Accepted: 03 April 2024
- Published online: 06 May 2024
Abstract: MicroRNAs (miRNAs) are non-coding, short regulatory molecules that govern plant development, stress responses, and environmental adaptations by targeting complementary RNA or DNA in a sequence-specific manner. The transcription and regulatory functions of miRNAs in plants are well understood, but we are just beginning to uncover the intricate regulatory activities of miRNAs in grasses. Here, we delve into the diverse regulatory roles of miRNAs and environmental sensing in grasses, encompassing species utilized for feed, forage, and ornamental purposes, such as switchgrass, ryegrass, bermudagrass, bluegrass, orchardgrass, and the model grass Brachypodium. We elucidate the biogenesis and regulatory cascade of miRNAs and explore the multifaceted developmental roles of miRNAs in grasses. Additionally, we provide insights into the crosstalk between miRNAs and their respective target mRNAs influencing the development of critical plant architecture. This review underscores the findings about biogenesis, targeting modes, and significant regulatory networks of miRNA to help illuminate key goals for future grass research.
-
Key words:
- miRNAs /
- Grasses /
- Endogenous cues /
- Exogenous cues /
- Post-transcriptional regulation












