-
Wheat (Triticum aestivum) is an important global food crop and contains carbohydrates along with other crucial nutrients such as proteins, small amounts of lipids, vitamins, minerals, as well as phytochemicals[1]. Dietary fiber is the carbohydrate oligomers and polymers, which are resistant to digestion and absorption in the human small intestine, leading to partially or complete fermentation in the human large intestine[2]. Whole wheat grains contain between 9% and 20% dietary fiber, and the main components of dietary fiber are cell wall polysaccharides, primarily arabinoxylan and (1,3;1,4)-β-D-glucan (β-glucan), representing approximately 70% and 20% of total dietary fiber, respectively[3]. In addition, dietary fiber in wheat grain also contains resistant starch, which is not digested in the small intestine and is able to reach the large intestine and colon relatively unchanged[4].
Wheat grains contain a starch-rich endosperm, an embryo (germ) and a fiber-rich outer covering (bran). Wheat-derived foods are made from whole wheat or white flour, which is made from wheat grains that have had the bran and germ removed[5]. Regular consumption of whole grain products has been advocated as a dietary recommendation, but white flour-based products, which are produced through milling to separate the bran and germ from the white flour, continue to dominate in most countries. Improving the fiber content of white flour preparations, and enhancing the general palatability of whole wheat preparations have the potential to improve fiber consumption.
Consumption of wheat-derived foods with high contents of arabinoxylan, β-glucan, and resistant starch can reduce the negative effects of diseases such as type 2 diabetes, as well as cardiovascular and gastrointestinal diseases[6−8]. Moreover, dietary intervention is increasingly encouraged as an effective way for preventing obesity and diabetes and is associated with beneficial effects on human health. Clinical analyses have demonstrated that dietary fiber exerts a beneficial impact on blood pressure and serum cholesterol levels, while also having a protective effect against the development of specific types of cancer, particularly colorectal and breast cancers[9].
In this review, we summarize research on three major components of dietary fiber in wheat: arabinoxylan, β-glucan, and resistant starch. We describe their compositions, biosynthetic pathways, and key enzymes, as well as the effects of these components on the health benefits of wheat fiber. The current advances in QTL/gene mapping and genetic improvement of wheat fiber are summarized and the challenges and perspectives on further research aimed at improving dietary fiber in wheat are discussed.
-
Arabinoxylan is a major component in the cell wall of wheat grain. In wheat starchy endosperm, arabinoxylan accounts for 70% of the cell wall polysaccharides and its content based on whole wheat bran ranges from 5% to 27%[10]. Arabinoxylan is one of the major sources of dietary fiber in human diet and is also a crucial factor determining wheat end-use quality and its utilization in animal feed and distilling[11]. The intake of arabinoxylan-rich bread is negatively correlated with postprandial glycemic responses in healthy adult subjects[12]. Indeed, in the gastrointestinal tract, arabinoxylan is intertwined with carbohydrates such as starch, thus delaying the absorption of carbohydrates and reducing post-meal blood sugar levels.
Composition and biosynthesis of arabinoxylan in wheat
-
The classification of arabinoxylan typically involves two fractions: water-extractable (WE-AX) and water-unextractable (WU-AX). WE-AXs constitute ~25%–30% of the total arabinoxylans in wheat grain[13]. The arabinoxylan in white flour consists of a linear (1,4) linked backbone of β-D-xylopyranosyl (xylose) residues[14]. The monomers that constitute the arabinoxylan polysaccharide are xylose and arabinose, which are biosynthesized in the cytoplasm and Golgi because of the existence of many isoforms of enzymes associated with biosynthesis. The arabinoxylan backbone is biosynthesized in the Golgi through the complex interplay of metabolites and the cytosol, that is a process regulated by various membrane channels including uridine diphosphate-sugar transporters.
Biosynthesis of the final arabinoxylan structure involves participation of two major types of enzymes including the glycosyltransferases (GTs) and glycoside hydrolases (GHs). GTs catalyze the development of glycosidic bonds through transferring the nucleoside diphosphate sugars, which act as a sugar donor containing a nucleoside phosphate, onto a particular receptor. Among the 124 GTs which are related to the biosynthesis of the cell wall in the wheat starchy endosperm, the genes encoding TaGT47_2, TaGT43_2, and TaGT43_1 involved in arabinoxylan backbone biosynthesis and TaGT61_1 in the arabinosylation of xylan are expressed at the highest levels[15−17] (Fig. 1). Specifically, all three homoeologs encoding TaGT43_2 and TaGT47_2 are expressed in the wheat endosperm and are regarded as a participant in the arabinoxylan backbone biosynthesis, while two xylan arabinosyl transferases, TaXAT1 and TaXAT2, belonging to the GT61 family, are involved in decorating the arabinoxylan backbone with arabinosyl and xylosyl sidechains[16]. Finally, members of the GH family could be involved in the breakdown of cellulose, allose, mannan, or arabinoxylan[15].
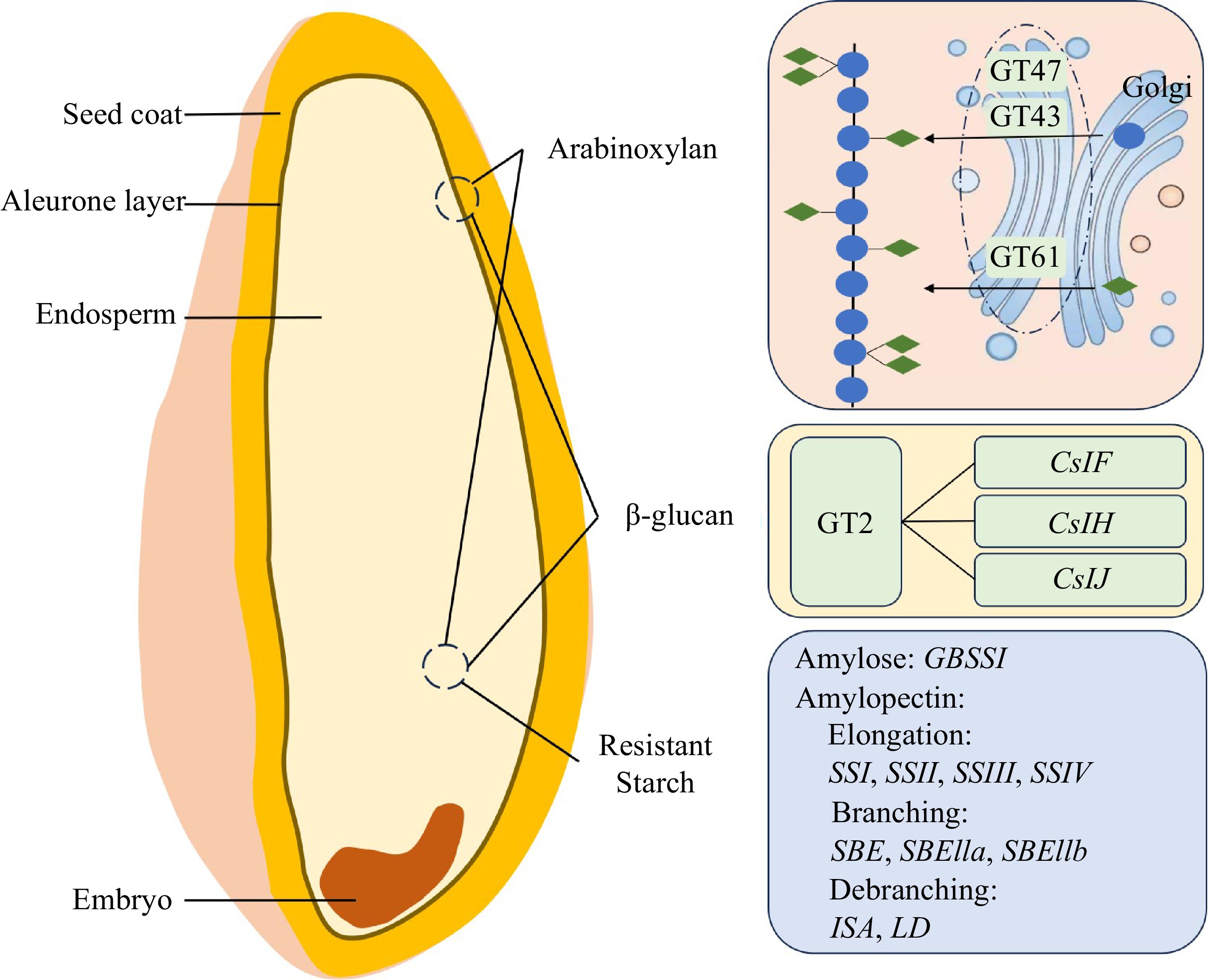
Figure 1.
The composition, biosynthetic pathways, and key enzymes for the three major classes of dietary fiber in wheat. GT47, GT43, GT61, and GT2 are glycosyltransferases. CslF, CslH, and CslJ are Cellulose-synthase-like (Csl) genes. GBSSI, granule-bound starch synthase I. SSI to SSIV, starch synthase I to IV. SBE, starch-branching enzyme. ISA, isoamylase-type debranching enzyme. LD, limit dextrinase. The blue ellipses represent UDP-D-xylose, and the green diamonds represent UDP- arabinopyranose.
Identification of QTLs/genes and development of molecular markers for arabinoxylan content in wheat
-
In recent years, multiple QTLs for arabinoxylan and WE-AX content have been identified using different wheat populations[18−20] (Table 1). Genes for arabinoxylan and WE-AX content have also been identified. Four genes were reported to participate in the arabinoxylan biosynthesis pathway; these genes encode a glucosyltransferase, a peroxidase, a glucosidase, and a methyltransferase[21]. Marcotuli et al. found 19 QTLs associated with arabinoxylan content[22], nine of which coincide with annotated genes encoding enzymes involved in arabinoxylan biosynthesis[15,17,23]. These genes include: i) Gal7, encoding the glycosyl hydrolase GH35; ii) a cluster of GT1 genes, including TaUGT1 and cisZog1; iii) CelC, encoding the glycosyl hydrolase GH1; iv) Ugt12887 and TaUGT1, both encoding the glucuronosyltransferase GT1; and v) Gsl12 and Cel8, encoding a (1,3)-β-D-glucan synthase and GH, respectively[22]. Eight candidate genes for arabinoxylan content such as genes encoding F-box domain proteins, the disease-resistance protein RPM1, and the transcription factor bZIP29 were identified through analysis of arabinoxylan content in 562 wheat genotypes combined with genome-wide association study (GWAS)[24]. QTLs with stable effects on total arabinoxylan or WE-AX content might be useful for improving arabinoxylan contents in wheat breeding through molecular marker-assisted selection. For example, QTL on chromosome 1BL of the Chinese wheat cultivar Yumai 34, contributed 24.2% of the phenotypic variation in arabinoxylan levels, leading to the molecular marker development based on allele-specific single nucleotide polymorphism (SNP)[25]. In addition, five SNPs on chromosomes 1BL and 5BS were reported to be associated with total arabinoxylan levels and 13 SNPs on chromosomes 1BL, 2BS, 6BS, 7A, and 7BL were related with WE-AX content[26]. Among which, SNP on 1BL were relevant to both traits and explained 13.29%–17.22% phenotypic variation for total arabinoxylan and 11.56%–19.37% for WE-AX, respectively and competitive allele-specific PCR (KASP) markers were developed for utilization during wheat breeding process.
Table 1. QTLs/genes and molecular markers for wheat dietary fiber content.
Trait QTL Chr Nearest marker Position (cM) Candidate genes Reference Arabinoxylan content WE-AX QAxvs.inra-1B 1B Xcfa2147 82.7 − [18] QAxfg1.inra-6B 6B wPt-2218 22.2 PC1 TOT-AX QGax.aww-2A.1 2A wpt-3114-2A 85.7 − [19] QGax.aww-4D.1 4D gpw-95001-4D 46.0 WE-AX QgWE-AX.caas-1B 1B HVM23–Sec1 − − [20] QgWE-AX.caas-5A 5A Xgwm443–Xcwem44 QgWE-AX.caas-5B 5B Xbarc142–Xwmc28 QgWE-AX.caas-7A 7A Xbarc174–Xbarc108 Grain dietary fiber content − − − − glucosyl-transferase (GT)
peroxidase glucosidase methyltransferase[21] Arabinoxylan content QGax.mgb-1A.1 1A wsnp_Ex_c45880_51550172 70.1 Gal7 [22] QGax.mgb-2B.1 2B Tdurum_contig45838_263 107.39 TaUGT1 QGax.mgb-3A.1 3A Kukri_c17966_634 122.68 CelC QGax.mgb-5A.1 5A Ex_c95453_1499 26.51 Ugt12887 QGax.mgb-5A.3 5A tplb0056b09_1000 63.69 TaUGT1 QGax.mgb-7A.1 7A Tdurum_contig69003_459 42.08 Gsl12 QGax.mgb-7A.2 7A wsnp_Ex_c21854_31021668 130.27 Cel8 Arabinoxylan content AX-95086356 1B − − F-box/FBD/LRR-repeat protein; [24] AX-94470319 4B Disease resistance protein RPM1/PIK6-NP-like; AX-94713015 5D bZIP transcription factor 29 Arabinoxylan content Y34Ukr-RH13-TOTAX; 1B − − − [25] Y34Cla-JI15-TOTAX Arabinoxylan content − 1B 1B_646895451 − − [26] 1B_653086336 1B_653681771 1B_654915479 5B 5B_14665450 2B 2B_184634480 6B 6B_26597224 7A 7A_234827309 7A_264333614 7A_458678969 7A_474572231 7A_516508921 7A_700824770 7B 7B_454100716 β-Glucan content QGbg.mgb-1A.1 1A Kukri_rep_c110838_253 10.6 − [35] QGbg.mgb-1A.2 1A Kukri_c43410_348 81.6 Cel9 QGbg.mgb-2A.1 2A Tdurum_contig10785_816 11.2 WSs2A QGbg.mgb-2A.2 2A Excalibur_c44834_80 197 Bamy1 QGbg.mgb-2B 2B BobWhite_c25359_132 14.5 Wxl1 QGbg.mgb-3B 3B BS00091867_51 97.2 Xip-II QGbg.mgb-5B 5B Tdurum_contig35470_227I 166 − QGbg.mgb-7A.1 7A tplb0024a09_829 49.7 − QGbg.mgb-7A.2 7A Tdurum_contig19352_76 90.9 1-FEH QGbg.mgb-2A.1 2A IWB1280 35.8–48.0 − [36] QGbg.mgb-2B.1 2B IWB30115 0.1–3.9 − QGbg.mgb-2B.2 2B IWB23783 29.9–47.9 GLU1a QBgn 3A Xwmc202–Xgwm2 [38] 1B Xwmc419–Xwmc134 5B Xwmc149–Xgwm335 6D Xhbe404–Xcfd188 QTL 1 4M 100022501_F_0 − glutathione S-transferase 3-like [39] QTL 2 5M 100013840_F_1 − QTL 3 1M 100079925_F_0 − Resistant starch content A-type starch granules Qga.caas-4AL 4A Xwmc262 – Xj133 − − [61] Qga.caas-1DL 1D Xcfd48.1 – Xcfd48.2 Qga.caas-7BL 7B Xwmc311 – Xbarc50 B-type starch granules − 4S TR132- TR126 − − [62] B-type starch granules − 4A − − BGC1-A [63] 4B
4DBGC1-B
BGC1-D4B
4DBGC1-B
BGC1-DAmylose content qhams7A.1 7A C7A.8493170 – C7A.39576499 40.0 GBSSI [65] Resistant starch content − 2D Xbarc59 − − [67] 5B Amylose and resistant starch content − 2ABD − − SBEIIa [50,70,71] 2ABD SBEIIb Amylose content − 7ABD − − SSIIIa [72−74] 1ABD SSIIIa Genetic improvement of arabinoxylan content in wheat
-
The genes which are responsible for the extension of xylan backbone in wheat are the putative orthologs of the Arabidopsis (Arabidopsis thaliana) GT genes IRREGULAR XYLEM10 (IRX10), IRX14, and IRX9, named TaGT47_2, TaGT43_1, and TaGT43_2, respectively[15]. Downregulating TaGT47_2 or TaGT43_2 expression by RNA interference (RNAi) resulted in as much as a 50% decrease in total arabinoxylan levels with a concomitant increase in arabinosylation levels ranging from 25% to 30%[17]. In these lines, the cell walls of starchy endosperm showed a reduction of xylan and arabinoxylan epitopes (as detected by immunolabeling) and a 50% reduction in cell wall thickness than the wild-type control. Freeman et al. also reported the reduction of WE-AX levels in wheat through downregulating TaGT43_2 and TaGT47_2[27]. A mutant with TaGT43_2 three homoeologs knock-out showed a reduction in arabinoxylan content of approximately 65%[28]. Silencing of the wheat TaXAT1 gene in the GT61 family, which is expressed at the highest levels in starchy endosperm, induced a 70%–80% reduction in α-(1,3)-linked arabinofuranosyl (Araf) residues substitution of β-(1,4)-D-xylopyranosyl (Xylp) residues in arabinoxylan[16]. Overall, these results provide evidence for the relevance of specific genes in controlling the arabinoxylan content in wheat. However, thus far, RNAi has not yet been used to improve arabinoxylan content.
-
β-glucan accounts for approximately 20% of total cell wall polysaccharides in wheat grain and is the second most abundant cell wall polysaccharide[10]. β-glucan is effective at reducing postprandial blood glucose levels[29]. β-glucans interact with lipids and biliary salts by forming glucan–biliary salt complexes directly or by creating a viscous layer that hinders absorption, thus decreasing the adsorption of biliary salts in the bowel[30], thereby reducing blood cholesterol levels and mitigating the risk of cardiovascular disease[7]. The β-glucan level in wheat grains is much lower relative to barley or oat. Particularly, the β-glucan content is 2.5%-11.5% in barley and 2%-7.8% in oats, but only 0.4%-1.4% in wheat[31]. The widespread consumption of wheat makes it an ideal target for enhancing β-glucan content.
Composition and key enzymes involved in β-glucan biosynthesis in wheat
-
β-glucans are long or short-chain polymers of glucose subunits connected by (1,3) and (1,4) linkages. Single (1,3) linkages are isolated by two or three (1,4) linkages, leading to trisaccharide and tetrasaccharide products after digestion by lichenase, which is a specific enzyme[32]. β-glucans are enriched in the subaleurone layer of the wheat seed, with a minor amount in the endosperm[33]. In cereals, β-glucans are biosynthesized by enzymes belonging to the cellulose synthase-like superfamily, which in turn is a component of the large glycosyltransferase GT2 family[34].
QTLs/genes associated with β-glucan content in wheat
-
Studies examining genetic variability associated with wheat β-glucan content using different wheat populations have been carried out in recent years (Table 1). Seven QTLs related to wheat grain β-glucan were found in tetraploid durum wheat, leading to the identification of candidate genes encoding GT or GH[35]. These genes include: i) Cel9, encoding the endo-β-(1,4)-glucanase GH9; ii) WSs2A, encoding the starch synthase II; iii) Bamy1, encoding the β-amylase GH14; iv) Wxl1, encoding the (1,4)-beta-xylanase GH10; v) Xip-II, encoding the xylanase inhibitor protein GH18; vi) 1-FEH, encoding the fructan 1-exohydrolase GH32. Three additional QTLs, named QGbg.mgb-2A.1, QGbg.mgb-2B.1, and QGbg.mgb-2B.2, were identified from a durum wheat using recombinant inbred line (RIL) population derived from a cross between two elite wheat cultivars Duilio and Avonlea by the same research group[36]. GLU1a gene encoding β-glucosidase 1a (GLU1a) was identified as candidate gene for QGbg.mgb-2B.2, and this enzyme catalyzes the hydrolysis of the glycosidic linkage in glycosides to form of intramolecular hemiacetal and hemiketal and the corresponding free aglycon[37]. In hexaploidy wheat, the QTLs on chromosomes 3A, 1B, 5B, and 6D were identified to be associated with β-glucan level[38] , among which the QTL located between markers Xwmc149 and Xgwm213 on chromosome 5B explained 15% of the phenotypic variation. Finally, Ivanizs et al. found three QTLs associated with β-glucan content in Aegilops biuncialis[39]: QTLs 2 and 3 were located on chromosomes 1M and 5M, and tetraploid wheat also have QTLs influencing the synthesis of β-glucan on group 1 and 5 chromosomes[40]. Another QTL 1 co-located with GTS3L encoding glutathione S-transferase participated in multiple metabolic pathways. However, most of these QTLs are not stable across different environments[36,38], which limited the utilization of marker-assisted selection during wheat breeding for β-glucan improvement. There are three cellulose-synthase-like (Csl) family members: CslF, CslH, and CslJ[41] (Fig. 1), among which CslF primarily participates in the synthesis of β-glucan in wheat. TaCslF6 has been identified as the crucial candidate for wheat β-glucan biosynthesis[42], with three homoeologs mapped near the centromeres on wheat chromosomes 7AL, 7BL and 7DL[32]. In hexaploid wheat, RNAi was employed to specifically suppress TaCslF6 in grain, leading to a 30% to 52% decrease in β-glucan content[42]. In durum wheat, Marcotuli et al. reported that the expression of CslF6 has a positive correlation with β-glucan content[43].
Genetic improvement of β-glucan in wheat
-
The strategies of interspecific hybridization and chromosome engineering can be used to introgress genes of distantly related wild and cultivated relatives to increase β-glucan content in wheat. Barley (Hordeum vulgare) has been successfully utilized in interspecific hybridization, since barley grain contained the highest β-glucan content[44]. It has been reported that HvCslF4, HvCslF6 and HvCslF9 located on chromosome 2HS, 7HL and 1HS greatly regulate β-glucan content in barley grain[45]. Several genes from barley were introgressed into wheat, which led to increased β-glucan content in wheat grain. Türkösi et al. developed a wheat/barley Robertsonian translocation (RobT) line 7BS.7HL, which confers increased grain β-glucan content (0.9%) than the control (0.7%)[46]. Danilova et al. developed a set of six compensating RobT chromosomes containing barley chromosome 7H (carrying the HvCslF6 gene related to β-glucan synthesis) and three group-7 chromosomes of wheat, among which the average β-glucan content of 7AS·7HL (1.01%), 7BS·7HL (0.82%), and 7DS·7HL (0.85%) were increased compared to the control (0.72%), although they were still much lower than the barley parent (5.81%)[32]. Subsequently, the same group used induced wheat–barley homologous recombination to shorten the barley chromosome 7HL arm in these RobTs to small segments[44]. They showed that the β-glucan level was increased in wheat lines containing two or four copies of the barley Cellulose synthase-like F6 gene (HvCslF6). Hence, by increasing the copies of the HvCslF6 gene through combining different recombinant chromosomes in a single line, the β-glucan content in wheat grain could be further increased. In addition, a wheat addition line containing the HvCslF9 gene showed a much higher β-glucan content[47]. Taken together, it is helpful to introgress barley HvCslF6 to wheat for β-glucan content improvement in wheat grains, but the β-glucan content in barley was still much more than that in translocation lines. Thus, other key genes from barley could be the candidates for increasing β-glucan biosynthesis in wheat.
For the purpose of identification of genotypes with high β-glucan content that can be utilized as interspecific gene resource transferring to cultivated wheat, Marcotuli et al. screened a set of cultivated and wild Triticum and Aegilops species for β-glucan content. They found two Aegilops species including Ae. umbellulata, UU genome and Ae. markgrafii, CC genome exhibiting superior β-glucan content than Triticum species and they were the putative candidates for wheat β-glucan content improvement[48]. Specifically, species with the U genome seemed to have a greatly higher β-glucan level (about 5.3%) compared to cultivated hexaploid bread wheat (about 0.83%). Another candidate gene is ISA1, encoding Isoamylase 1 (ISA1), which is an isoamylase-type debranching enzyme playing an important role in amylopectin biosynthesis. Sestili et al. knocked down Isa1 in durum wheat using RNAi: the resulting transgenic lines had reduced starch content, as well as moderately enhanced phytoglycogen and β-glucan contents[49]. These findings demonstrate that transgenic strategy can be utilized to regulate the content of β-glucan in wheat grains.
-
Starch is the main polysaccharide in wheat, which can be classified into amylose and amylopectin based on the polymerization form of glucose[50]. Amylose, a linear structure connected by α1, 4 bonds, is densely packed within the starch granule in the wheat grain endosperm[50]. Amylopectin, a branched molecule containing both α1, 4 and α1, 6 bonds, generates the complex structure that occupies most of space within the starch granule of wheat grain endosperm. In wheat grain, amylopectin typically constitutes over 70% of the total content, while amylose accounts for less than 30%. According to its digestibility, starch is generally categorized as rapidly and slowly digestible starch, or resistant starch, depending on its post-consumption behavior[51]. Resistant starch refers to dietary starch that cannot be digested in the small intestine and undergoes fermentation exclusively in the colon[52]. The consumption of resistant starch has been shown to directly attenuate postprandial blood glucose levels, serum cholesterol levels, the glycemic index, and insulin levels, thereby contributing to the prevention or reduction of obesity, diabetes, cardiovascular disease, and chronic kidney disease[8]. There are five types of resistant starch (type 1−5), the augmentation of amylose content can decelerate the rate of starch digestion and increase the content of resistant starch type 2[53]. By focusing on biochemical pathways that enhance the amylose content within a starch granule allows for the synthesis of high-amylose grains with higher levels of resistant starch type 2, and consequently an increased amount of dietary fiber in refined flour[53].
Biosynthesis of resistant starch in wheat
-
Starch biosynthesis is a complicated and finely-regulated process that requires several functional enzymes, including sucrose synthase (SUS), ADP-glucose pyrophosphorylase (AGPase), granule-bound starch synthase I (GBSSI), starch synthase (SS), starch-branching enzyme (SBE) and de-branching enzyme (DBE)[54,55] (Fig. 1). In this process, the sucrose biosynthesized in leaves is transported into grains, and then it is decomposed into fructose and UDP-glucose by SUS. Following this, the products are converted into ADP-glucose by AGPase. Subsequently, soluble ADP-glucose is transported into the amyloplasts as a direct substrate, where the starch biosynthesis is initiated[8]. GBSSI controls amylose biosynthesis independently, while SBE, DBE and SS are responsible for amylopectin biosynthesis[54−56]. The amount of resistant starch is positively related to the proportion of amylose[57]. Therefore, increasing GBSSI or decreasing SS and SBE gene expression is the most common means to increase the resistant starch content in cereal endosperm.
QTLs/genes and molecular markers for resistant starch content in wheat
-
In recent years, multiple QTLs and genes related to resistant starch content in wheat have been reported (Table 1). The quantity and proportion of A- or B-type starch granules, as well as the quantity of amylose, are related to resistant starch content. A-type granules contain higher amounts of amylose than B-type starch granules[58]. The proportion of B-type granules is positively related to amylose and resistant starch content[59,60]. Using 240 RILs derived from a cross between wheat lines PH82-2 and Neixiang 188, three QTLs associated with the contents of A-type starch granules, named Qga.caas-1DL, Qga.caas-7BL, and Qga.caas-4AL, were identified, explaining 5.6%, 5.2%, and 3.8% of the phenotypic variation, respectively[61]. Howard et al. developed an F2 population of Aegilops with variation in B-type starch granule content and used this population to identify one major QTL accounting for 44% of the phenotypic variation[62]. Similarly, the progeny derived from Ae. peregrina × KU37 were crossed, revealing a link between B-GRANULE CONTENT 1 (BGC1) and resistant starch content[63]. BGC1 inhibits the formation of A-granules during early grain development but promotes the formation of B-granules during middle grain development.
In general, starch with a greater proportion of amylose has a higher level of resistant starch[64]. Thus, QTLs and genes regulating amylose content might also affect resistant starch content. Analysis of an F2 population developed by crossing the high-amylose donor 'TAC 75' and high-yielding variety 'WH 1105' identified qhams7A.1, a stable QTL associated with amylose content. This QTL includes the GBSSI gene[65]. A high-amylose line carrying the missense mutation g.35767184 T > C in GBSSI produces the mutant protein GBSSI_L539P, which is associated with high-amylose starch. The L539P mutation improves GBSSI activity by affecting its starch-binding ability, suggesting that this mutant allele could be a molecular target for enhancing amylose and resistant starch content in wheat grain[66]. Various additional molecular markers related to resistant starch content have also been developed. For example, bulk segregation analysis identified the SSR marker Xbarc59 located on chromosomes 2D and 5B of spring wheat[67]. In the F2 generation obtained from a cross between Wuchun 4 and M344, genotypes with high, medium, and low resistant starch contents, were efficiently screened using the Xbarc59 marker.
Ethyl methanesulfonate (EMS)-targeted mutagenesis has been successfully used to develop wheat lines with high amylose and high resistant starch contents. In common wheat grains, the amylose content varies from 20%–30%. Mishra et al. produced a bread wheat population comprising 101 EMS-induced mutant lines, with amylose content ranging from ~3%–76% and resistant starch content ranging from 1–41%[68]. Irshad et al. identified stable mutants with significant changes in resistant starch content using an EMS-induced population of wheat variety J411[69].
Four types of enzymes are required for starch biosynthesis: GBSSI is required for amylose biosynthesis, while amylopectin is produced by the concerted action of several classes of starch synthase (SSI, SSII, SSIII, and SSIV), branching (SBEI, SBEIIa, and SBEIIb), and debranching enzymes[54,55]. By suppressing the expression of SBE or SS genes or enhancing the expression of the GBSSI gene in the amyloplast, starch biosynthesis can be modified to increase the amylose content, thus leading to an increased proportion of resistant starch.
GBSSI
-
The waxy (Wx) gene encodes GBSSI. A mutation in Wx-B1 reduced GBSSI activity by affecting its starch-binding ability, indicating that modifying GBSSI activity can modulate amylose content. Accordingly, screening an EMS-mutagenized population of bread wheat variety J411 revealed two mutants with high levels of resistant starch and two mutants with low levels of resistant starch, with the mutants with high levels of resistant starch showing higher GBSSI expression and the mutants with low levels of resistant starch showing higher SBE expression[69].
SBEII
-
Durum wheat with a SBEIIa/b-AB double mutation exhibited a 22% and 115% increase in amylose and resistant starch content, respectively[70]. Introducing a series of deletions and SNPs into SBEIIa and SBEIIb also increased the amylose content to an unprecedented level of ~85%, with a concomitant increase in resistant starch content[50]. Schönhofen et al. detected an increase in amylose content of up to 60% and a 10-fold increase in resistant starch content in SBEII mutants[71]. The combination of Targeting Induced Local Lesions in Genomes (TILLING) technology and breeding led to the successful introduction of SBEIIa mutations in bread wheat and durum wheat[49]. These mutants showed increases in amylose and resistant starch contents ranging from 47%–52.7% and 4.7%–6.8%, respectively.
SSII and SSIII
-
Whole meal flour from a SSIIIa triple mutant had more resistant starch and higher levels of arabinoxylans and β-glucans than the wild type[72]. Schoen et al. identified SSIIa knockout mutations in all three genomes of the wheat variety 'Jagger', generating a triple knockout mutant affecting the activities of all three homoeologs[73]. The triple knockout mutant had increased amylose and resistant starch levels without the shriveled grain phenotype observed in other SSIIa knockout mutants[74−76]. Importantly, since all the mutants were obtained using an elite wheat cultivar, they are immediately available for practical application.
Genetic improvement of resistant starch content in wheat
-
Plant breeding using crop varieties with high levels of resistant starch and marker-assisted selection are effective methods for improving resistant starch content. Combined mutations in SBEIIa and SBEIIb paralogs by EMS were obtained in hexaploid wheat cultivar Lassik including: SBEIIa/b-AB; SBEIIa/b-A, SBEIIa-D; SBEIIa/b-B, SBEIIa-D; and SBEIIa/b-AB, SBEIIa-D. The quintuple mutant line (SBEIIa/b-AB, SBEIIa-D) exhibited a significant increase in amylose and resistant starch contents[77]. Hogg et al. also observed an increase in total fiber content in pasta made from a SSIIa null durum wheat line[75]. In addition, Botticella et al. developed a breeding strategy to produce a set of durum wheat SSIIa null mutants[78]. The authors introgressed SSIIa null mutations into the Italian elite durum wheat cultivar Svevo and showed that the semolina flour produced from two SSIIa null mutant lines had increased arabinoxylans, β-glucans, and resistant starch contents. The same authors also used mutagenesis followed by conventional marker-assisted breeding to generate three mutant lines that produced starch with an amylose content of 0%, 46%, and 79%[76].
RNAi has also been used to generate transgenic wheat lines with increased amylose content. Downregulating SBEIIb alone had no effect on amylose content, whereas the concomitant downregulation of SBEIIa and SBEIIb resulted in starch comprising more than 70% amylose[79]. When rats were fed whole meal flour from the RNAi double downregulated lines, improvements in several indices of large-bowel function were observed. Similarly, the RNAi-mediated downregulation of SBEIIa in durum wheat induced an up to 75.05% increase in amylose content[80]. Finally, suppressing SSI by RNAi dramatically enhanced amylose content (31.4%)[81].
Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated nuclease 9 (Cas9) is widely used to introduce specific DNA modifications into targeted genes[82]. This method has been successfully used to create transgene-free lines with various amylose contents in crops including rice and maize[83−85]. Using the modern winter wheat cv. Zhengmai 7698 and the spring wheat cv. Bobwhite, Li et al. employed CRISPR/Cas9-targeted mutagenesis of TaSBEIIa to successfully generate transgene-free high-amylose plants[86]. The flours of lines affected in the functions of all three homoeologs showed increased amylose, resistant starch, protein, and soluble pentosan contents, which benefit human health.
-
The physicochemical properties of arabinoxylan, β-glucan, and resistant starch are closely related to dietary fiber content in wheat. Further optimization of arabinoxylan, β-glucan, and resistant starch biosynthesis in wheat endosperm represents an important future direction in wheat breeding for improving the nutritional quality of grain. Nonetheless, there are still many hurdles to overcome.
Improving dietary fiber content in wheat negatively affects grain yield
-
The increased amylose and resistant starch contents in SBEII, SSII, and SSIII mutants are related to changes in grain characteristics that may limit their usefulness in breeding programs. Indeed, these mutants show reduced starch contents and grain weight, leading to yield penalties[70−76]. For example, Hazard et al. reported that mutations in the durum wheat genes SBEIIa/b-AB were associated with an average reduction in kernel weight of 5.2% and a 15% reduction in grain yield[70]. Schönhofen et al. found that SBEII mutants exhibited reductions in kernel weight and grain yield of 2.8% and 5.8%, respectively[71]. Schoen et al. reported that SSIIa triple mutants had a 21.29% reduction in thousand-grain weight, which was also significantly reduced in SSIIa single mutants compared to the control[73]. Similarly, targeting of SBEIIa by CRISPR/Cas9 was associated with a slight reduction in grain length and width, resulting in a decrease in the thousand-grain weight in different mutant lines, with the highest effect observed in aabbdd triple-null lines[86]. Thus, the effects of partial- or triple-null alleles are dosage dependent, with triple-null lines exhibiting the most profound impacts on yield penalty. Accordingly, Fahy et al. reported that the SSIIIa triple mutants exhibited shrunken grains with a weight reduction of ~11%, whereas the grains of SSIIIa single and double mutants were not significantly different from those of the control[72]. The same research group also revealed a possible dosage effect on starch content. Marcotuli et al. reported QTLs associated with wheat grain β-glucan, while the associated genes include starch synthesis II, supporting a link between the biosynthetic pathways of starch and β-glucan[35], and the negative correlation between grain weight and β-glucan content were also observed in wheat[48].
Improving dietary fiber content in wheat negatively affects processing quality and consumer acceptability
-
High-amylose wheat varieties have been successfully developed during the past decades through modification of the SBEII, SSII, or SSIII genes, which lead to a reduced glycemic index and are beneficial for human health[70,72,75]. Unfortunately, the grain- or flour-processing quality of these varieties was negatively affected. Given the crucial role of a continuous gluten network, the dilution effect, mechanical shear effect, competitive water absorption, and steric hindrance effect of dietary fiber may potentially disrupt structure, thereby resulting in suboptimal rheological properties of dough[53]. For example, Schönhofen et al. reported that SBEII mutations exert notable influences on bread-making quality. The traits related to end-use quality such as enhanced grain hardness, and increased starch damage, water absorption, and protein content in flour; with diminished flour extraction, farinograph development, stability times, starch viscosity, and loaf volume[71]. Li et al. found that increasing amylose content drastically reduced cooking quality and that bread made from high-amylose wheat flour had a lower loaf volume, a more dense crumb structure, and higher hardness than wild-type wheat bread[64]. Li et al. reported that bread and biscuits made of high-amylose flours from TasbeIIa triple-null lines had decreased volume, brighter color, higher hardness, and lower sensory scores compared to the wild type[86].
Durum wheat SBEIIa/b-AB mutant lines showed favorable performance on pasta firmness but negative effects on pasta color and semolina extraction[70]. These mutants showed an average adverse in firmness of 12.4% relative to the wild-type lines. However, the average amount of semolina extracted from grains after milling was 4.6% lower in the SBEIIa/b-AB mutants vs the wild type. The SBEIIa/b-AB mutants also displayed an average increase in cooking loss of 20.4%, a 6.1% reduction in cooked weight, and a 10.4% decrease in color score. Furthermore, Hogg et al. reported that durum wheat accessions containing SSIIa null mutation showed high-amylose content and lead to increased grain protein content with lower semolina yield[75]. Pasta made from semolina of the SSIIa null mutant exhibited reduced water absorption, higher cooking loss, shorter cooking time, and significantly firmer texture even when overcooked, as compared to the wild-type line. The brightness of cooked and uncooked pasta from the SSIIa null mutant was diminished compared to the wild type. Therefore, efforts aimed at improving dietary fiber content must monitor effects on processing quality and select for traits with minimal impacts on quality traits. Consequently, the balance of grain protein, starch and dietary fiber is more important, enhancing the dietary fiber content of wheat while preserving yield and end-use quality.
Improving wheat dietary fiber by increasing bran or aleurone layer thickness
-
The wheat endosperm is composed of an outer layer called bran (14%−15%), an inner starchy endosperm (81%−83%) and embryo (2%−3%). The bran, composed of the aleurone, endocarp, mesocarp, and epicarp, forms the outer layer of the grain. It contains trace minerals and indigestible cellulose materials, including the β-glucans. In contrast to barley, the distribution of β-glucans in the wheat endosperm is less consistent and at lower levels. The aleurone layer was found to have high concentrations of β-glucans, accounting for approximately 29% of its dry weight[42]. As a dietary fiber supplement incorporated into food, wheat bran exhibits remarkable efficacy in promoting cardiovascular and gastrointestinal health[87]. Therefore, increasing bran or aleurone layer thickness is an effective way to improve dietary fiber in wheat. Actually, in rice, Li et al. & Liu et al. has proved that increasing the thickness of aleurone layer is an effective way to improve the nutrition properties. They screened the ta1 and ta2 mutants displaying increase in the number of aleurone cell layers compared to the wild type, resulting in elevated levels of all examined nutritional factors, including lipids, proteins, vitamins, minerals, and dietary fibers[88,89]. The ta1 aleurone thickened phenotype is caused by mutations in the OsmtSSB1, which interacts with RECA3 and TWINKLE to inhibit abnormal recombination of mitochondrial genomic DNA in rice aleurone cells, ensuring optimal mitochondrial energy supply[88].The phenotype of ta2 is attributed to a dominant negative mutation in the DNA demethylase gene OsROS1, which holds a pivotal role in epigenetics based on molecular genetic evidence. In wild-type rice cells, this mutation leads to a notable increase in the aleurone layer from 1−2 to 4−10 layers, consequently enhancing the nutritional quality of rice caryopsis significantly[89]. In wheat, Chen et al. reported a lgp1 mutant with low gluten protein exhibiting significantly increased bran content and dietary fiber content compared to the wild type[90], which could be used for wheat dietary fiber improvement.
Improving wheat dietary fiber content by fine-tuning the expression of starch biosynthesis-related genes
-
As discussed above, partial-null and triple-null alleles of starch biosynthesis-related genes, especially SSIIa, SSIIIa, and SBEII triple-null lines, incur yield penalties and affect wheat processing quality. In particular, triple-null lines demonstrated more profound impacts on yield penalty and end-use quality than other mutants. This observation suggests that fine-tuning the expression of genes or the activities of enzymes involved in amylose content could be an effective strategy to increase fiber content while minimizing impacts on grain weight and end-use quality. For example, it might be possible to use unique combinations of mutant ssIIa, ssIIIa, or sbeIIa homoeologs in wheat breeding programs. In addition, moderate changes in the expression of starch biosynthesis-related genes that are highly expressed in the endosperm could improve dietary fiber content without severely affect yield and quality traits in rice[91,92]. Specifically, Xu et al. generated a series of Wxb mutants using cytidine base editors (CBEs)[92], leading to 1.4%–11.9% increases in amylose content without affecting the quality or appearance of milled rice. Another approach could be to target starch biosynthesis-related genes with relatively low expression levels in endosperm, since mutations in these genes tend to cause moderate changes in starch biosynthesis, thus limiting undesired effects[93].
Several other studies successfully fine-tuned gene expression using CRISPR/Cas9 to target gene regulatory sequences[85,94]. Huang et al. generated six novel Wx alleles by editing the region near the TATA box of the Wxb promoter[91]. This resulted in downregulated Wx expression and increased grain amylose content. Liu et al. engineered quantitative variation for yield-related traits in maize by generating weak promoter alleles of CLAVATA3/embryo surrounding region-related (CLE) genes and a null allele of a newly identified and partially redundant compensating CLE[95]. While CLE knockout alleles had a deleterious effect on yield, fine-tuning CLE expression by editing its cis-regulatory elements boosted yield. Finally, CRISPR-dCas9 can be utilized to target the addition or removal of DNA methylation to silence or activate genes in Arabidopsis[96]. Therefore, modulating DNA methylation at cis-regulatory elements represents an additional strategy to fine-tune the expression of genes.
For β-glucan improvement, the identification of the key functional genes such as TaCslF6 had been contributed to increased β-glucan content[43]. However, the lack of major and stable QTL still limited and prevented the utilization of marker-assisted selection for β-glucan improvement in wheat breeding. Thus, enhancing our understanding on the genetic regulation of β-glucan biosynthesis and identifying more superior genetic resources for enhancing β-glucan content is essential. The integration of high-throughput markers with genome sequencing techniques holds promise for genetic identification, elucidating genetic control, and developing molecular markers linked to key genes and QTL associated with this trait[87−89].
As discussed above, different (and non-mutually exclusive) strategies can be adopted to fine-tune gene expression and/or enzymatic activity. We provided several examples in which these strategies were efficiently employed in rice, maize, and Arabidopsis. In Fig. 2, we propose several strategies for improving dietary fiber content in wheat grain. However, it remains to be assessed whether and to what extent these approaches can be used to increase dietary fiber content in wheat without affecting yield and end-use quality. This is one of the major challenges that should be addressed in the future.
This work was supported by the National Natural Science Foundation of China (Grant No. 32125030), the National Key Research and Development Program of China (2022YFD1200203), and Frontiers Science Center for Molecular Design Breeding (Grant No. 2022TC149).
-
The authors confirm contribution to the paper as follows: study conception and design: Yao Y, Yang C, Chen Q; data collection: Yang C, Chen Q, Zhang X, Zhang J; draft manuscript preparation: Yang C, Chen Q; manuscript revision: Rossi V, Du J, Xin M, Ni Z, Sun Q, Yao Y. All authors have reviewed and approved the final version of the manuscript.
-
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
-
The authors declare that they have no conflict of interest.
- Copyright: © 2024 by the author(s). Published by Maximum Academic Press on behalf of Hainan Yazhou Bay Seed Laboratory. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Yang C, Chen Q, Zhang X, Zhang J, Rossi V, et al. 2024. Genetic improvement of dietary fiber in wheat grains. Seed Biology 3: e002 doi: 10.48130/seedbio-0024-0002
Genetic improvement of dietary fiber in wheat grains
- Received: 09 October 2023
- Revised: 10 January 2024
- Accepted: 22 January 2024
- Published online: 06 February 2024
Abstract: Dietary fiber has multiple health benefits, including reducing blood glucose levels and enhancing gut health. Wheat (Triticum aestivum), the most important staple crop in the world, provides a good source of dietary fiber. Thus, improving dietary fiber composition and content of wheat grain is a critical task for wheat research and breeding programs. The main components of dietary fiber in wheat grain include arabinoxylan, β-glucan, and resistant starch. This review summarizes recent advances in research on wheat dietary fiber and their impacts on human health. We discuss quantitative trait locus/gene mapping and genetic approaches for improving dietary fiber in wheat, as well as challenges and prospects in this research area.
-
Key words:
- Dietary fiber /
- Wheat grain /
- Arabinoxylan /
- β-glucan /
- Resistant starch