-
The genus Diorygma (family Graphidaceae) includes lichenized fungi having ecorticate to pseudocorticate thallus, resulting in a matte, farinose upper surface; lirellate ascocarp with a whitish pruinose disc; an uncarbonized to occasionally carbonized, narrow exciple; clear, I+ blue hymenium; often anastomosing and laterally branched paraphyses; distinct epithecium; 1–8 spored unitunicate clavate asci; mostly muriform to transversely septate, hyaline, very rarely brownish ascospores; and the presence of norstictic, stictic, and/or protocetraric acid thallus chemistry. Asexual morph not reported[1,2].
The Diorygma was first introduced by Eschweiler in 1824, with Fries[3] comparing it to Coniangium (Arthonia). Later, Massalongo[4] accepted Diorygma as a valid genus but did not describe any new species under it. Staiger[5] revisited the taxonomy and reclassified species into the genera Fissurina and Platythecium. Meanwhile, Müller[6] described Graphina as a genus characterized by muriform spores, and despite recognizing the uniqueness of Diorygma, Awasthi & Joshi[7] introduced the name Cyclographina, overlooking the earlier and validly published name Diorygma. Staiger[5] later reintroduced Diorygma but proposed placing it outside Graphidaceae. However, Kalb et al.[1] revived and monographed the genus, documenting 31 species and providing molecular phylogenetic evidence (based on nuLSU) to confirm its position within Graphidaceae, and is currently kept under the same family[8]. Further molecular studies, including those by Rivas Plata et al.[9] (using mtSSU, nuLSU, and RPB2), highlighted the paraphyletic nature of Diorygma and Thalloloma lineages, a finding corroborated by Ansil et al.[10].
A total of 86 of Diorygma have been reported globally, of which 43 species have been recorded from India[10−16]. Notably, 29 species occur in the Western Ghats, highlighting the importance of this lichen-rich biodiversity hotspot. Despite the documented diversity within the genus, only 24 taxa currently have sequence data available in GenBank, and among these, only 17 have been identified to the species level. This limited molecular sequence representation underscores a significant gap in the phylogenetic characterization of Diorygma species, particularly given the reports of overlapping morphological characters within the genus[10], and between the genus Thalloloma[9]. Such overlap complicates species delimitation and highlights the need for comprehensive DNA sequencing to support accurate taxonomy, resolve cryptic diversity, and enhance phylogenetic understanding.
The present study forms part of an ongoing project to unravel the symbiotic relationships within the lichen family Graphidaceae from the Western Ghats (WG), aiming at identifying and delimiting species of Diorygma and their photobionts using traditional taxonomic methods and multimarker (mtSSU, nuLSU, and RPB2) molecular data to assess the diversity of these lichens from diverse habitats of the WG, India.
-
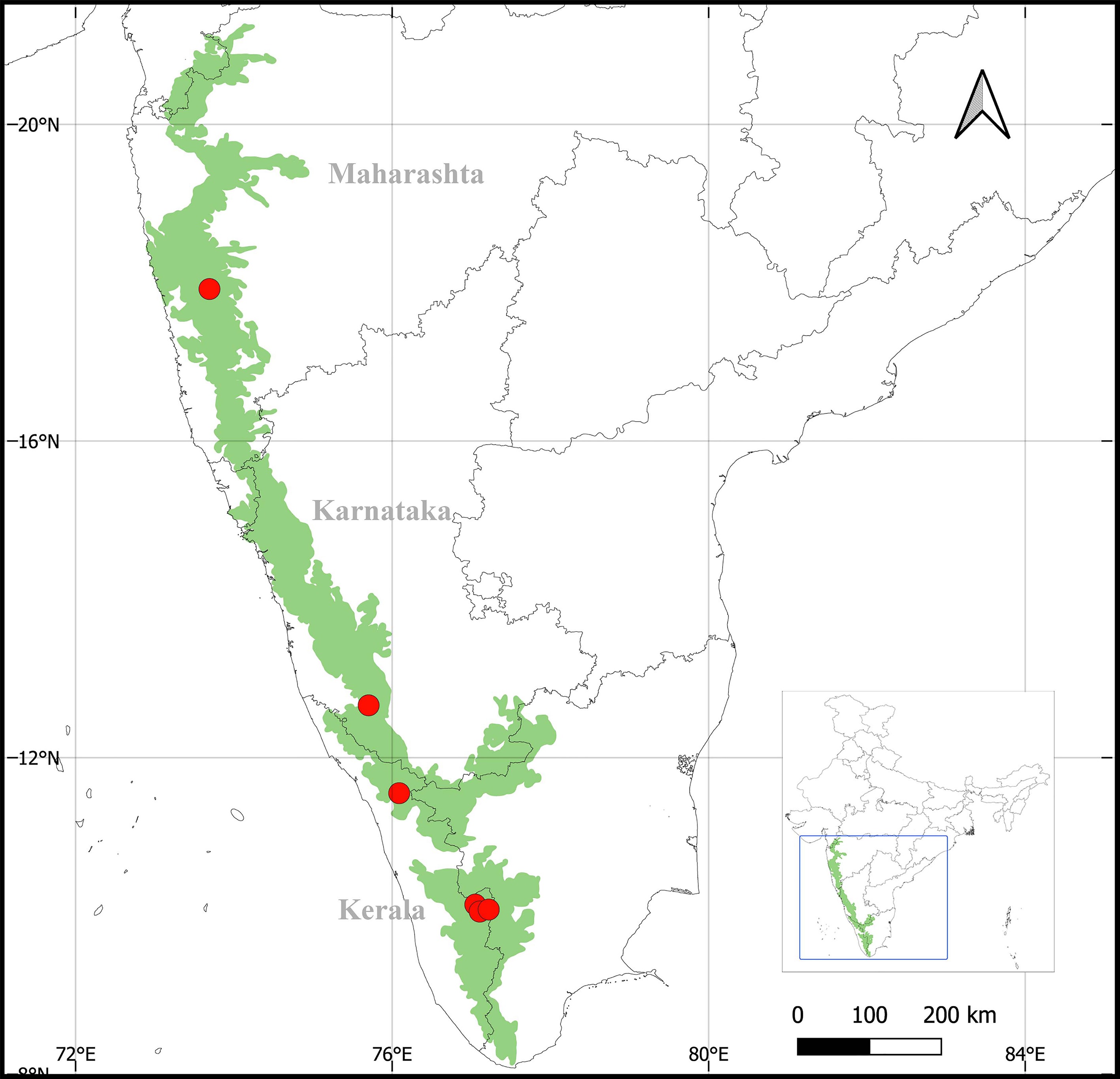
The sampling of Diorygma lichen thalli was carried out during 2022–2023 from the Southern WG, including Chembra Peak in Wayanad district and Devikulam, Eravikulam, Kolukkumala of the Idukki district in Kerala state; the Central WG, including Pushpagiri of the Kodagu district in Karnataka state; and the Northern WG, including Lingmala in the Satara district in Maharashtra state of India (Fig. 1). The collections were made from an altitude range of 1,122–2,139 msl, annual rainfall 1,468–2,642 mm, and average annual temperature 24.4–25.7 °C.
Morphological and chemical analyses
-
Morphology was examined with a binocular stereomicroscope (Olympus SZX16 with a DP23 camera, Japan). Longitudinal sectioning of lirellae was carried out using a razor blade, followed by mounting in lactic acid (with gentle flame heating), 10% potassium hydroxide (K), water, Lugol's iodine (I), and KI (Potassium hydroxide, followed by Lugol's iodine) separately for microscopy. The microscopic measurements were taken by mounting the sections in water. Microscopic observations were made using an Olympus BX53 with a DP74 camera (Japan). Morphological identification followed pertinent taxonomic references[1,17]. Thallus chemical profiles were examined by thin layer chromatography (TLC) following standard lichen microchemical protocols[18], with the solvent systems toluene : 1,4-dioxane : acetic acid (TDA, 180:45:5), and toluene : ethyl acetate : formic acid (TEF, 139:83:8). The studied specimens were stored at the Ajrekar Mycological Herbarium (AMH) at MACS Agharkar Research Institute, Pune, India.
DNA extraction, polymerase chain reaction, and sequencing
-
Extraction of DNA and amplification of gene regions were performed using the Sigma RED Extract-N-AmpTM Seed PCR Kit, according to the manufacturer's protocol, in a ProFlexTM PCR system thermocycler (Applied Biosystems, USA). CHtrente2.for[19], and ITS4T[20] were primers for amplifying ITS markers from the photobiont. For the mycobiont, primers for amplification were: i) for the mtSSU marker[21], mrSSU1 and mrSSU3R; ii) for the LSU marker, AL2R[22], and LR6[23]; iii) for the RPB2 marker, GD1-RPB2-7cF and GD-RPB2-11aR[24]. PCR conditions for amplification were: 5 min initial denaturation at 95 °C, followed by 30 cycles for 1 min at 94 °C and 35 cycles for 30 s at 55 °C (ITS), 35 cycles for 1 min at 50 °C (mtSSU), 35 cycles for 1 min at 58 °C (nuLSU), 35 cycles for 1 min from 57 to 72 °C, with 1 °C increase in temperature per cycle (RPB2), and final extension for 10 min at 72 °C. The PCR products were purified with the Alphagen PCR Purification Kit (Alphagen Biotech Ltd., Ping Tung, Taiwan, China), and sequenced using the same primers with the BigDye Terminator v. 3.1 Cycle Sequencing Kit, and the sequencing reactions were run on an ABI PRISM® 3100 Genetic Analyzer (Applied Biosystems, USA).
Phylogenetic analyses
-
The nucleotide database of NCBI GenBank was searched using MegaBLAST[25] for the nearest matching Diorygma and Trentepohlia sequences. An updated phylogeny of Diorygma was assembled following relevant phylogenetic references[10,15], with the addition of other sequences (mtSSU, nuLSU, and RPB2) of the genus from GenBank (Table 1). The Trentepohlia phylogeny was assessed following relevant references[19,26] with the additions of available ITS sequences retrieved from GenBank. Newly generated DNA sequences in this study were deposited in GenBank (Table 2).
Table 1. Species name, voucher information, and accession numbers for the mycobiont sequences used in this study.
Name of taxa Specimen voucher Country mtSSU nuLSU RPB2 Diorygma aeolum AMH22.09 India PV031879 PV034271 − D. aeolum AMH22.13 India PV031880 PV034272 − D. aeolum AMH22.257 India PV031869 PV034274 PV038023 D. aeolum AMH22.259 India PV031870 PV034273 PV038022 D. aeolum AMH23.486 India PV031871 PV034276 − D. aeolum AMH23.488 India PV031872 PV034275 − D. aeolum AMH23.502 India PV031873 − − D. aeolum AMH23.526 India PV031876 − − D. aeolum AMH23.583 India PV031877 − − D. aeolum AMH23.612 India PV031878 − − D. aeolum AMH23.637 India PV031874 − − D. aeolum AMH23.659 India PV031875 − − D. antillarum E:Nelsen 4037 USA JX046452 JX046465 − D. antillarum E:Nelsen 4185 Brazil JX046451 JX046464 − D. antillarum E:Lücking 33019 El Salvador JX046454 JX046467 − D. antillarum E:Lücking 33018 El Salvador JX046453 − − D. circumfusum Herb. Kalb 33922 Australia DQ431963 AY640019 − D. erythrellum Kalb 38772 Thailand JX421022 − − D. hieroglyphicum V. & R. Wirth No. 26647 French Polynesia − AY640015 − D. junghuhnii MSSRF/Dj/G23/2015 India MN944822 − − D. junghuhnii Herb. Kalb 33254 Brazil − AY640016 − D. junghuhnii Herb. Kalb 33931 Australia − AY640017 − D. junghuhnii Herb. Kalb 33937 Australia DQ431962 − − D. junghuhnii MSSRF/Dj/Mycobiont India MN944821 − − D. junghuhnii Lumbsch 20539l Fiji JX421023 JX421474 − D. junghuhnii Herb. Kalb 33937 Australia − AY640018 − D. karnatakense AMH21.26 India OP235521 OP235516 OP245173 D. karnatakense AMH21.52 India OP235522 OP235517 OP245174 D. karnatakense AMH21.54 India OP235523 OP235518 OP245175 D. karnatakense AMH21.55 India OP235524 OP235519 OP245176 D. karnatakense AMH21.60 India OP235525 OP235520 OP245177 D. microsporum Lücking 26504 USA JX421024 − − D. minisporum Lumbsch 19543v Kenya HQ639598 HQ639626 − D. poitaei Lücking 28538 Nicaragua HQ639596 HQ639627 − D. poitaei Lücking 28533 NIcaragua JX421025 JX421475 − D. pruinosum Mangold 28g Australia − JX421476 − D. pruinosum Herb. Kalb 26578 Australia − AY640014 − D. pruinosum Herb. Kalb 26612 Australia DQ431964 − − D. sipmanii F. Berger No. 14011 Costa Rica DQ431961 AY640020 − D. tiantaiense Jia ZJ19123 China − MW750692 − Diorygma sp. Lumbsch 20501la Fiji − JX421478 − Diorygma sp. Lumbsch 20513a Fiji − JX421477 − Diorygma sp. Lumbsch 19082l Australia − JX421479 − Thalloloma anguinum RivasPlata 2063 Philippines JX421337 − − T. anguinum Lumbsch 19804c Fiji JX421336 − − T. hypoleptum Rivas Plata 17570 Philippines JF828970 − − T. hypoleptum Rivas Plata 17573b Philippines HQ639609 − − Phaeographis intricans Kalb 38864 Thailand JX421254 JX421602 JX420924 Newly generated sequences are given in bold. Table 2. Species name, voucher information, and accession numbers for the algal sequences used in this study.
Name of taxa Voucher/isolate/strain information Country ITS Printzina cf. lagenifera TreFl 137 Isolate TreFl 137 United Kingdom JQ617961 Printzina lagenifera TreFl 13 Isolate TreFl 13 Austria JF727811 Printzina lagenifera TreFl 14 Isolate TreFl 14 Italy JF727814 Printzina lagenifera TreFl 143 Isolate TreFl 143 United Kingdom JQ617954 Printzina lagenifera TreFl 30 Isolate TreFl 30 Austria JQ617955 Trentepohlia abietina GD1352 Isolate GD1352 China KX586810 Trentepohlia annulata SAG_20.94 Isolate SAG_20.94 Czehia JQ687378 Trentepohlia annulata SY1318 Isolate SY1318 China KX586840 Trentepohlia arborum fTTW Isolate fTTW Thailand JX675738 Trentepohlia arborum TreFl31 Isolate TreFl31 Brazil KC489136 Trentepohlia arborum FJ1321 Isolate FJ1321 China KX586802 Trentepohlia aurea TreFl38 Isolate TreFl38 Argentina KC489138 Trentepohlia aurea TreFl39 Isolate TreFl39 Argentina KC489139 Trentepohlia aurea DZ1320 Isolate DZ1320 China KX586791 Trentepohlia cf. arborum HZ-2017 Isolate FJ1320 China KX586801 Trentepohlia cf. arborum HZ-2017 Isolate YN1047 China KX586847 Trentepohlia cf. aurea HZ-2017 Isolate YN1240 China KX586858 Trentepohlia cf. jolithus HZ-2017 Isolate DZ1317 China KX586788 Trentepohlia cf. umbrina HZ-2017 Isolate GD1350 China KX586813 Trentepohlia cf. umbrina HZ-2017 Isolate GX1306 China KX586815 Trentepohlia dialepta SY1321 Isolate SY1321 China KX586841 Trentepohlia diffusa YN1262 Isolate YN1262 China KX586861 Trentepohlia jolithus ASIB505 Isolate ASIB505 Austria JX675737 Trentepohlia jolithus YJG Isolate YJG China KX586842 Trentepohlia minima YN1234 Isolate YN1234 China KX586856 Trentepohlia prolifera YN1243 Isolate YN1243 China KX586859 Trentepohlia rigidula FJ1302 Isolate FJ1302 China KX586793 Trentepohlia rigidula SY1310A Isolate SY1310A China KX586838 Trentepohlia sp. AMH22.244 Voucher AMH22.244 India PP002593 Trentepohlia sp. AMH22.257 Voucher AMH22.257 India PV031732 Trentepohlia sp. AMH22.259 Voucher AMH22.259 India PV031731 Trentepohlia sp. HZ-2017 Isolate DZ1318A China KX586789 Trentepohlia sp. HZ-2017 Isolate FJ1305 China KX586794 Trentepohlia sp. HZ-2017 Isolate FJ1313B China KX586797 Trentepohlia sp. HZ-2017 Isolate FJ1323B China KX586803 Trentepohlia sp. HZ-2017 Isolate FJ1324B China KX586804 Trentepohlia sp. HZ-2017 Isolate GX1304 China KX586814 Trentepohlia sp. HZ-2017 Isolate GX1319 China KX586818 Trentepohlia sp. HZ-2017 Isolate GX1326 China KX586819 Trentepohlia sp. HZ-2017 Isolate GX1332 China KX586824 Trentepohlia sp. HZ-2017 Isolate GX1343 China KX586828 Trentepohlia sp. HZ-2017 Isolate GX1345 China KX586829 Trentepohlia sp. HZ-2017 Isolate SY1302 China KX586836 Trentepohlia sp. HZ-2017 Isolate SY1305 China KX586837 Trentepohlia sp. HZ-2017 Isolate SY1314B China KX586839 Trentepohlia sp. HZ-2017 Isolate YN1206 China KX586852 Trentepohlia sp. HZ-2017 Isolate YN1235 China KX586857 Trentepohlia sp. HZ-2017 Isolate YN1316 China KX586862 Trentepohlia sp. HZ-2017 Isolate YN1317 China KX586863 Trentepohlia sp. HZ-2017 Isolate YN1034A China KX586777 Trentepohlia sp. HZ-2017a Isolate HN1008 China KX586834 Trentepohlia sp. KR-2023a Isolate RKCSP316RK05 India OR602551 Trentepohlia sp. S16_Gp Strain S16_Gp Argentina JF727817 Trentepohlia sp. TreFl 144 Isolate TreFl 144 United Kingdom JQ617980 Trentepohlia sp. TreFl 149 Isolate TreFl 149 United Kingdom JQ617963 Trentepohlia sp. TreFl163 Isolate TreFl163 Costa Rica KC489153 Trentepohlia sp. TreFl35 Isolate TreFl35 Argentina KC489110 Trentepohlia sp. TreFl51 Isolate TreFl51 Brazil KC489123 Trentepohlia sp. TreFl61 Isolate TreFl61 Brazil KC489122 Trentepohlia sp. TreFl63 Isolate TreFl63 Brazil JX675731 Trentepohlia sp. TreFl82 Isolate TreFl82 Brazil KC489142 Trentepohlia sp. TreFl96 Isolate TreFl96 Brazil KC489120 Trentepohlia umbrina fTCA Isolate fTCA Austria JX675736 Phycopeltis epiphyton YN1201 (IHB) Voucher YN1201 (IHB) China KP067278 Newly generated sequences are given in bold. A concatenated dataset of the mtSSU, nuLSU, and RPB2 sequences of the mycobiont and individual ITS dataset of the photobiont were analyzed, and conflicts between the trees generated from the individual markers of mycobionts were checked. The datasets were first individually aligned, edges trimmed, and edited manually in MEGA v. 11.0.11 using MUSCLE[27]. The alignment file was converted into PHYLIP format[28] using AliView v. 1.28. Marker partitioning was performed manually for the combined mtSSU + nuLSU + RPB2 dataset (nuLSU: 1–904 bp; RPB2: 905–1710 bp; mtSSU: 1,711–2,546 bp).
Phylogenetic analyses were carried out in IQ-TREE v. 2.1.3[29], and RAxML v. 8.1.11[30,31] using maximum likelihood (ML). Node support was estimated using 1,000 ultrafast bootstrapping (UFboot) in IQ-TREE and 1,000 rapid bootstrapping in RAxML. The selection of an appropriate model was done by the 'auto' option in IQ-TREE. In the combined mtSSU + nuLSU + RPB2 dataset, 'TNe + G4' was the best model for nuLSU, 'K2P' for mtSSU, and 'TIM + F + I + G4' for RPB2. Also, 'TN + F + I + G4' was found to be the best model for photobiont ITS. For RAxML analysis, 'GTR G + I' was determined a priori for individual and combined mtSSU + nuLSU + RPB2 and photobiont ITS datasets. For the selection of models for Bayesian posterior probability (PP) analysis, MrModeltest2 v. 2.4[32] was used according to the Akaike Information Criterion (AIC). MrBayes v. 3.2.7[33] was used to perform Bayesian posterior probability (PP) analysis. 'GTR + I + G' was the best model for combined mtSSU + nuLSU + RPB2 and photobiont ITS datasets. For phylogenetic trees generated, only clades with UFboot BS ≥ 95% were considered supported for trees obtained using IQ-TREE, and BS ≥ 70% were considered supported for RAxML. Estimation of PP was done allowing independent rate variation and unlinked parameter estimation. A variant of the Markov Chain Monte Carlo (MCMC) method was used for sampling of trees. In 30,00,000 generations, trees were sampled every 1,000th generation (resulting in 3,000 trees) after running four simultaneous Markov chains. The first 750 trees (25%) were discarded, containing the burn-in phase of the analyses. The calculation of posterior probabilities (PP) was done using the remaining 2,250 trees in the consensus tree. In the Bayesian framework, clades with PP ≥ 0.95 were considered supported. The resulting phylogenetic trees were visualized using FigTree 1.4.0.[34].
-
Diorygma aeolum (Stirt.) Pushpi Singh & Kr. P. Singh, The Lichenologist 49(5): 527 (2017) (Fig. 2)
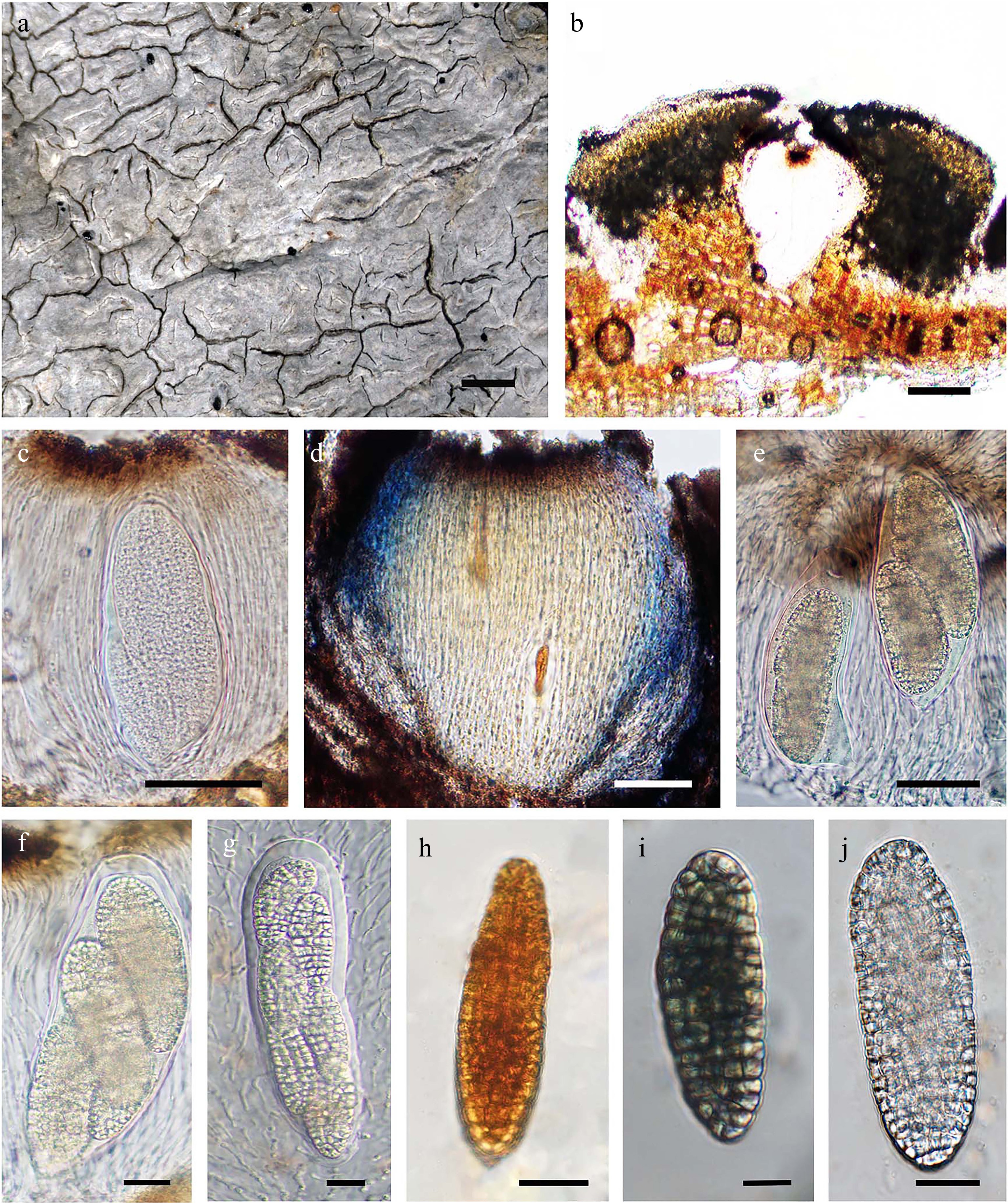
Figure 2.
Diorygma aeolum (AMH22.257). (a) Thallus surface. (b) Lateral section (LS) of lirellae. (c) Hymenium not-inspersed. (d) Hymenium laterally I+ blue in Lugol's iodine. (e)–(g) Asci with a varied number of ascospores. (h) I– ascospore. (i) I+ blue ascospore. (j) Ascospore in water. Scale bars: (a) = 1 mm, (b) = 100 μm, (c) = 10 μm, (d), (e) = 50 μm, (f)–(h) = 20 μm, (i), (j) = 10 μm.
Index Fungorum number: IF819997
≡ Graphis aeola Stirt., Proc. Roy. phil. Soc. Glasgow 11: 316 (1879)
= Graphina aeola (Stirt.) Zahlbr., Cat. Lich. Univers. 2: 394 (1923)
= Diorygma albovirescens Makhija, Chitale & B.O. Sharma, Mycotaxon 109: 382 (2009)
= Diorygma megasporum Kalb, Staiger & Elix, Symb. bot. upsal. 34(no. 1): 160 (2004)
= Diorygma saxicola B.O. Sharma & Makhija, Mycotaxon 109: 215 (2009)
Thallus crustose, corticolous, thick, whitish to pale grey or greenish grey, indistinct to distinctly pseudocorticate, surface matt, continuous to cracked, not delimited by hypothallus, crystals present; Apothecia lirellate, abundant, randomly distributed, immersed, straight to flexuous, simple to irregularly branched, 0.3–4.5 mm × 0.2–1 mm, edges acute; Disc concealed to slit-like, greyish white pruinose; margin concolorous to thallus, ±raised; Exciple poorly developed, uncarbonized, convergent to slightly divergent, entire to sometimes striate, apically brownish, yellowish laterally and towards base; Epithecium distinct, dark brown 13–18 µm high; Subhymenium hyaline, laterally I+ blue-violet, 10–20 µm high; Hymenium clear, 110–200 µm high, laterally KI– blue-violet; Paraphyses smooth, branched, brownish and anastomosed towards apex; Asci clavate, unitunicate; Ascospores (1–)2–6(–8) per ascus, hyaline, ellipsoidal, muriform with all locules of equal size, 65–190 µm × 20–50 µm, mostly I+ blue, rarely I–; Pycnidia not observed.
Chemistry: Thallus K–, UV–, TLC: stictic and constictic acids (major), some specimens contain cryptocystic and norstictic acid (trace).
Distribution: High altitude regions of India and Myanmar.
Materials examined: India: Maharashtra: Satara, Lingmala (17°55'15" N, 73°41'23" E), on tree bark, 1,296 msl 15 February 2022, Ansil P. A. and Rajeshkumar K. C. (AMH22.09, AMH22.13); Karnataka: Kodagu, Pushpagiri wildlife sanctuary (12°39'38" N, 75°42'09" E) on tree bark, 1,122 msl, 05 December 2023, Ansil P. A. and Rajeshkumar K. C. (AMH23.583, AMH23.612); Kerala: Idukki, Devikilam (10°03'16" N, 77°06'23" E) on tree bark, 1,536 msl, 18 December 2022, Ansil P. A. and Rajeshkumar K. C. (AMH22.257, AMH22.259); Idukki, Kolukkumala (10°04'51" N, 77°13'14" E) on tree bark, 2,139 msl, 26 October 2023, Ansil P. A. and Rajeshkumar K. C. (AMH23.486, AMH22.279, AMH23.502); Idukki, Eravikulam national park (10°08'37" N, 77°03'01" E) on tree bark, 1,758 msl, 27 October 2023, Ansil P. A. and Rajeshkumar K. C. (AMH23.526); Wayanad, Chembra peak (11°33'13" N, 76°05'02" E) on tree bark, 1,124 msl, 17 December 2023, Ansil P. A. and Rajeshkumar K. C. (AMH23.637, AMH23.659).
Notes: Examination of D. albovirescens type material showed its similarity with D. aeolum with respect to indistinct pseudocortex; simple to branched lirellae; pruinose disc; Clear hymenium; muriform, ascospore septation, size, and thallus chemistry (Fig. 3). The protologue of D. saxicola shows agreement in morphology, having indistinct pseudocortex; simple to branched lirellae; brown disc; laterally I+ve clear hymenium; ascospore size, septation, and thallus chemistry with that of D. aeolum (Fig. 4). Therefore, D. albovirescens and D. saxicola are synonymized under D. aeolum following the taxonomic priority. The spore size of D. aeolum (65–190 µm × 20–50 µm) is comparable to D. subalbatum (75–145 µm × 24–34 µm), but D. subalbatum has shorter lirellae, 1–8 spores per ascus, and chemistry characterized by the presence of additional norstictic acid along with stictic acids. Phylogenetically, D. aeolum forms a sister clade to D. karnatakense, which is comparable to D. aeolum in having simple to branched lirellae; pruinose disc; Clear, I+ blue, hymenium and muriform ascospores, but differs with respect to an ecorticate thallus, slightly larger ascospores (75–220 µm × 18.5–51.5 μm) and norstictic, salazinic acid chemistry.

Figure 3.
Diorygma albovirescens (holotype). (a), (b) Holotype herbarium material. (c), (d) Thallus. (e)–(j) Ascus showing ascospores. Scale bars: (c) = 1 mm, (d) = 500 μm, (e)–(j) = 20 μm.

Figure 4.
Diorygma saxicola (holotype). (a), (b) Holotype herbarium material. (c), (d) Thallus. (e)–(g) Ascus showing ascospores. Scale bars: (c) = 1 mm, (d) = 500 μm, (e), (f) = 100 μm, (g) = 20 μm.
Phylogenetic results
-
The closest hits based on the MegaBLAST search of the GenBank nucleotide database, using the sequences, are given in Table 3.
Table 3. Details of closest search results of sequenced regions based on MegaBlast.
Sequence Organism Country Accession no. Identities Gaps mtSSU Diorygma karnatakense voucher AMH21.54 India OP235523 719/736 (98%) 3/736 (0%) Diorygma antillarum isolate MPN528 El Salvador JX046453 686/731 (94%) 19/731 (2%) Diorygma poitaei isolate DNA3210 Nicaragua HQ639596 685/731 (94%) 23/731 (3%) nuLSU Diorygma sp. XZ-2021a voucher Jia ZJ19123 China MW750692 773/847 (91%) 12/847 (1%) Graphidaceae sp. ETRP-2011 voucher Lücking 19955d1 Thailand JF828975 751/861 (87%) 23/861 (2%) Dyplolabia afzelii voucher AMH22.119 India OR602550 750/860 (87%) 28/860 (3%) RPB2 Diorygma karnatakense voucher AMH21.55 India OP245176 749/799 (94%) 2/799 (0%) Diorygma minisporum isolate CHAR48 Kenya KF875520 665/798 (83%) 0/798 (0%) Diorygma aff. minisporum isolate P14636 South Africa ON492053 647/789 (82%) 0/789 (0%) ITS Trentepohlia sp. isolate TreFl81 Brazil KC489141 429/459 (93%) 11/459 (2%) Trentepohlia sp. isolate TreFl56 Argentina KC489140 434/478 (91%) 29/478 (6%) Trentepohlia sp. HZ-2017 isolate FJ1323B China KX586803 374/441 (85%) 24/441 (5%) The combined mtSSU + nuLSU + RPB2 sequence data of Diorygma aeolum were analyzed with other available sequences of the genus in GenBank to determine the species placement (Table 1, Fig. 5). The phylogenetic tree was rooted with Phaeographis intricans voucher Kalb 38864. The phylogenetic trees of the combined mtSSU + nuLSU + RPB2 alignments estimated with IQ-TREE, RAxML, and MrBayes were topologically similar and hence, the tree resulting from IQ-TREE is presented (Fig. 5), with support values from the RAxML and Bayesian posterior probability analysis superimposed.
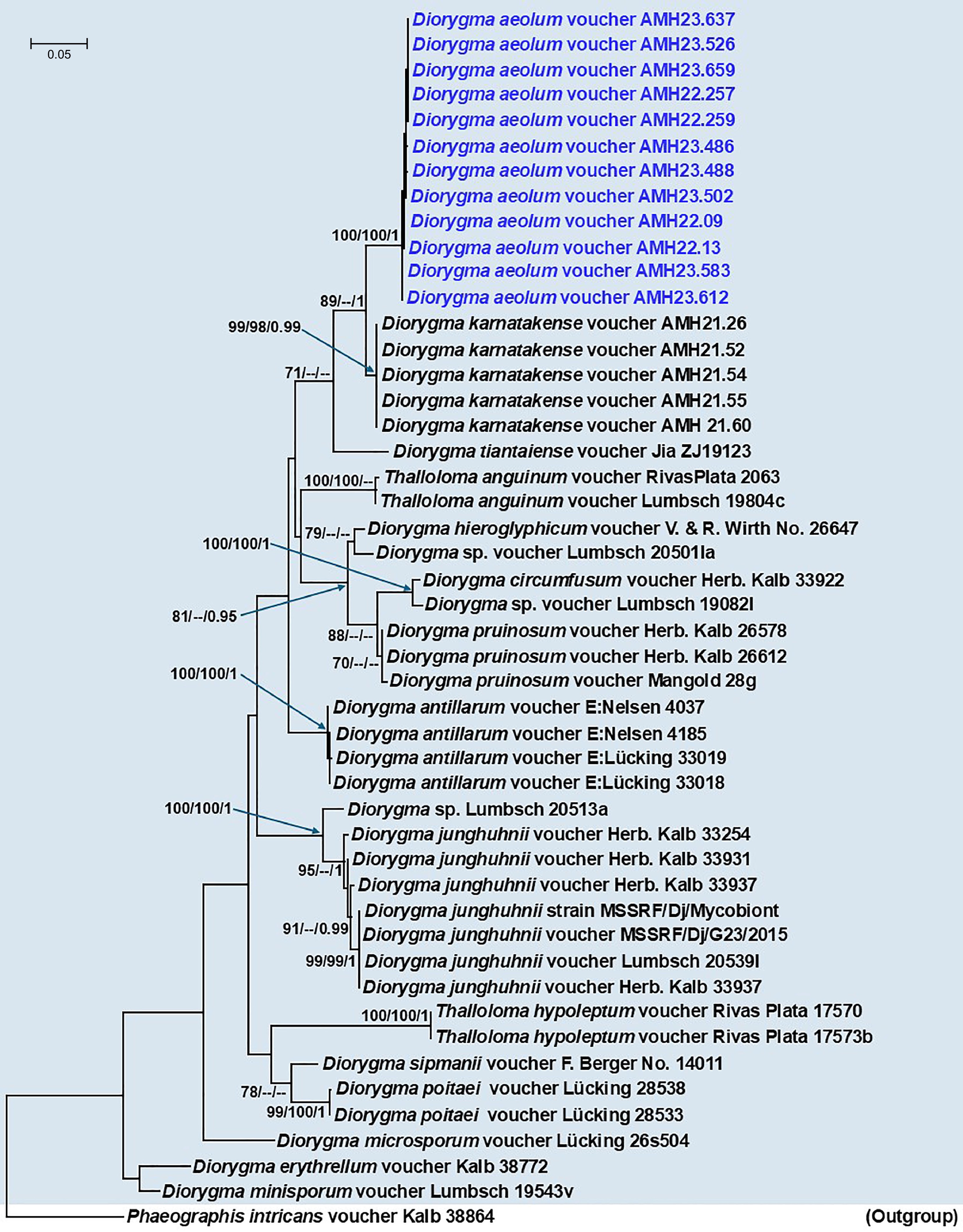
Figure 5.
Phylogenetic tree generated from Maximum Likelihood (ML) analyses based on the combined mtSSU + nuLSU + RPB2 data for the genera Diorygma and Thalloloma species (Graphidaceae). Node support from 1,000 non-parametric bootstraps for RAxML (R-BS), IQ-TREE (UFboot-BS), and posterior probability (PP) from MrBayes are shown at the nodes (R-BS ≥ 70%/Ufboot-BS ≥ 95%/PP ≥ 0.95). Low support values are not denoted. The tree is rooted with Phaeographis intricans voucher Kalb38864. The sequence generated for Diorygma aeolum in this study is indicated in blue.
In the phylogenetic tree, the sequences of the genera Diorygma and Thalloloma appear to be nested within each other, which Diorygma aeolum, newly sequenced in this study, formed a well-supported clade (89/--/1) sister to D. karnatakense (Fig. 5). The clade containing D. aeolum and D. karnatakense formed a less supported (71/--/--) clade allied to D. tiantaiense voucher Jia ZJ19123 reported from China[15].
The ITS sequences of Trentepohlia spp. were analysed with additional published sequences in the genus following pertinent phylogenetic references[19−26] to identify the species. The tree was rooted with Phycopeltis epiphyton voucher YN1201. The phylogenetic trees estimated with IQ-TREE, RAxML, and MrBayes were topologically similar and hence, the tree resulting from IQ-TREE is presented (Fig. 6), with support values from the RAxML and Bayesian posterior probability analysis superimposed. The results of the phylogenetic analyses are given in Table 4.
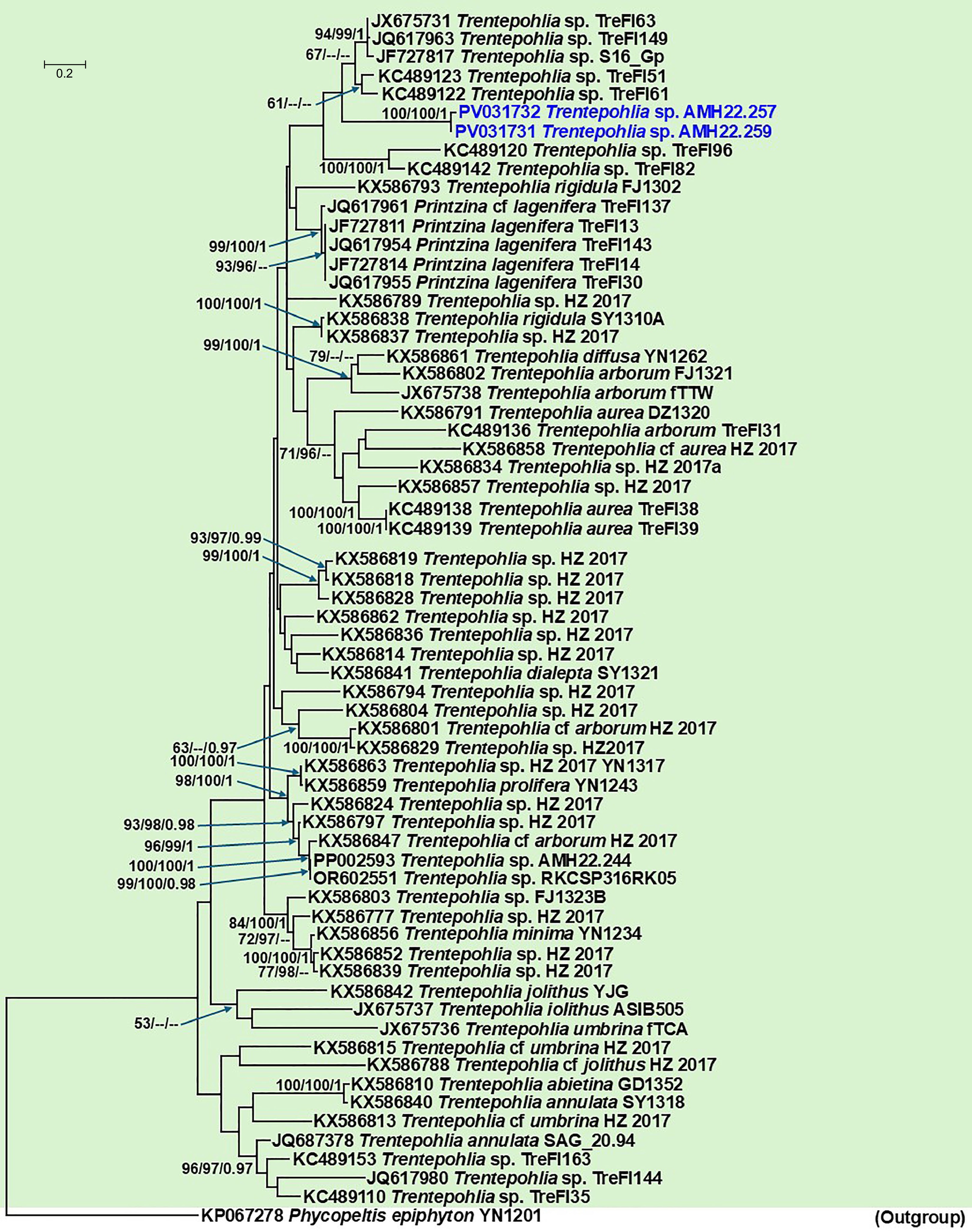
Figure 6.
Phylogenetic tree generated from Maximum Likelihood (ML) analyses based on the combined ITS data for the genus Trentepohlia (Trentepohliaceae), and allied Printzina species. Node support from 1,000 non-parametric bootstraps for RAxML (R-BS), IQ-TREE (UFboot-BS), and posterior probability (PP) from MrBayes are shown at the nodes (R-BS ≥ 70%/Ufboot-BS ≥ 95%/PP ≥ 0.95). Low support values are not denoted. The tree is rooted with Phycopeltis epiphyton voucher YN1201. The sequence generated for Trentepohlia sp. in this study is indicated in blue.
Table 4. Summary of results of the ML analyses performed.
mtSSU + nuLSU +
RPB2Photobiont
ITSNumber of strains (incl. outgroup) 48 65 Length incl. gaps 2,546 886 Distinct alignment patterns 470 675 Undetermined characters (gaps) 48.57% 31.05% Estimated base frequencies A: 0.296 A: 0.248 C: 0.183 C: 0.273 G: 0.255 G: 0.278 T: 0.266 T: 0.202 Substitution rates AC: 1.00000 AC: 1.00000 AG: 3.85637 AG: 1.36556 AT: 1.96997 AT: 1.00000 CG: 1.96997 CG: 1.00000 CT: 10.84645 CT: 2.98126 GT: 1.00000 GT: 1.00000 IQ-TREE analysis Model (BIC) K2P (mtSSU)
TNe+G4 (nuLSU)
TIM+F+I+G4 (RPB2)TN + F + I + G4 Final likelihood value −8,215.620 −14,901.181 Gamma distribution shape parameter (α) 0.155 0.759 RAxML analysis Model (BIC) GTR G + I GTR G + I Final likelihood value −8,246.352430 −14,886.765071 Gamma distribution shape parameter (α) 1.745954 0.784593 The Trentepohlia photobiont species sequenced from the studied specimen vouchers AMH22.257, and AMH22.259 formed a poorly supported clade allied to the clade containing Trentepohlia spp. isolates TreFl63 (from Graphis sp., Brazil), TreFl149 (from Arthonia atra, UK), S16_Gp (from Graphis propinqua, Argentina), TreFl51 (from Diorygma pruinosum, Brazil), and TreFl61 (from Graphis sp., Brazil) (Fig. 6). The position of this clade is yet to be defined because of the lack of significant statistical support. However, it forms an adjacent clade allied to Printzina lagenifera and a polyphyletic Trentepohlia rigidula. The Diorygma aeolum photobiont represents a distinct, unnamed species of Trentepohlia. The results of the phylogenetic analyses are given in Table 3.
-
This study represents a modern taxonomic approach for the characterization of Diorygma species and their photobiont Trentepohlia species from the Western Ghats of India.
Singh & Singh[17], while revisiting the type materials of Graphis aeola and Diorygma megasporum, proposed a new combination as D. aeolum for G. aeola and synonymized D. megasporum under D. aeolum. In the protologue, G. aeola was reported to have 1–4 spored asci, and similarly, D. megasporum with 2–6 spored asci. However, the description of Singh & Singh[17] was found to be facile, as the inconsistent mention of D. aeolum having 4–6 spored asci. In the present study, the collected specimens were found to have overlapping morphological characters and thallus chemistry with D. albovirescens and D. saxicola, hence, through revisiting the type materials, D. albovirescens and D. saxicola are synonymized under D. aeolum. In the present study, collected specimens were found to have overlapping ranges in ascospore numbers, such as 1–3, 1–4, and 1–6 spores per ascus in different samples. Regardless of this variation, the phylogeny inferred from the combined mtSSU + nuLSU + RPB2 tree defined the twelve accessions a one well-supported clade. According to Hawskworth[35], variations in spore number in an ascus can result from different reasons, with varying degrees of systematic importance, and the observed variation can be because of variations in the number of ascospores that mature within individual asci rather than more central systematic variations. Ansil et al.[10] also arrived at a similar conclusion regarding D. karnatakense, which is also collected from the Western Ghats and forms a clade sister to D. aeolum in the present study. Hence, the weightage of the difference in spore number as a species delimiting character in the genus Diorygma warrants further study incorporating morphological and molecular methods.
Lichens are established by associating with locally adapted photobionts[36], and the photobiont selectivity and specificity play a significant role in deciding lichen symbiotic outcomes[37]. Studies on photobionts in Graphidaceae are limited, making it difficult to place the Diorygma photobiont results in a broader perspective. In the preceding studies[19], photobiont strains belonging to Printzina lagenifera and Trentepohlia sp. were isolated from Graphis propinqua, G. scripta, and G. submarginata from Austria, Italy, and Argentina. In the genus Diorygma, Kosecka et al.[38] assessed the phylogenetic relationships with the Trentepohlia photobionts from Bolivia using rbcl2 data and explained 87% variation in the photobiont selectivity of Diorygma spp., 52% accounts for the mycobiont species, and 35% variability between species of mycobionts, climate, and geographic distance in combination. Borgato et al.[39] later substantiated the observation, reporting that certain photobiont lineages exhibit high specificity, associating exclusively with Diorygma antillarum, while others show broader associations with multiple Diorygma species. Photobiont studies involving Diorygma species from the Western Ghats have not yet been conducted. However, recent studies[10,40] on Allographa effusosoredica and Dyplolabia afzelii, both collected from the Western Ghats, revealed the presence of a shared Trentepohlia photobiont, suggesting a possible trend of locally adapted photobionts in the region.
The evolutionary predictions in Trentepohlia species of lichenized fungi are still in their infancy. Species such as Trentepohlia arborum and T. rigidula were found to be paraphyletic in nature. The presence of Printzina species, closely related to Trentepohlia spp., also requires future investigation. The Trentepohlia sp. sequenced in this study are found allied to the clade containing other yet to be identified Trentepohlia spp. sequenced from lichen genera such as Diorygma, Graphis, and Arthonia. Future research on the molecular phylogeny of both the mycobiont and photobiont, coupled with spatiotemporal analysis and host variations, is essential for a deeper comprehension of the variation of photobionts within Diorygma from the Western Ghats.
Kunhiraman C. Rajeshkumar and Bharati Sharma express gratitude to SERB, Department of Science and Technology, Government of India, for financial support (project CRG/2020/000668), and the Director, MACS Agharkar Research Institute, Pune, for providing the laboratory facilities. Parayelil A Ansil thanks CSIR-HRDG, India, for financial support under the SRF fellowship (09/670(0093)/2021-EMR-I). The authors thank the Kerala, Karnataka, and Maharashtra forest departments for permission and the logistics for sample collection. SERB, Department of Science and Technology, Government of India, (project CRG/2020/000668), and the CSIR-HRDG, India SRF fellowship (09/670(0093)/2021-EMR-I).
-
The authors confirm contributions to the paper as follows: study conception and design: Ansil PA, Rajeshkumar KC; data collection: Ansil PA, Rajeshkumar KC, Gaikwad S; analysis and interpretation of results: Ansil PA, Rajeshkumar KC, Lücking R, Sharma B; draft manuscript preparation: Ansil PA, Rajeshkumar KC, Gaikwad S, Lücking R, Sharma B. All authors reviewed the results and approved the final version of the manuscript.
-
All data generated or analyzed during this study are included in this published article and its supplementary information files. The alignment files used for phylogenetic analysis are made available in FigShare: (https://figshare.com/articles/dataset/Diorygma_aeolum_Concat_alignment/28309292).
-
The authors declare that they have no conflict of interest.
- Copyright: © 2025 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Ansil PA, Rajeshkumar KC, Lücking R, Gaikwad S, Sharma B. 2025. Resolving the phylogeny of Diorygma aeolum, along with its photobiont Trentepohlia species from the Western Ghats, India. Studies in Fungi 10: e023 doi: 10.48130/sif-0025-0022
Resolving the phylogeny of Diorygma aeolum, along with its photobiont Trentepohlia species from the Western Ghats, India
- Received: 09 June 2025
- Revised: 12 August 2025
- Accepted: 28 August 2025
- Published online: 24 October 2025
Abstract: This study unravels the symbionts of the lichenized fungus Diorygma aeolum with its Trentepohlia photobiont from the Western Ghats (India). Phylogenetic positioning of the mycobiont and photobiont was assessed by integrating morphology, thallus chemistry, and DNA sequence data. In the phylogenetic analysis of the mycobiont using mtSSU, nuLSU, and RPB2 sequences, D. aeolum was placed in a clade closely related to D. karnatakense. Based on the study, the species D. albovirescens and D. saxicola under D. aeolum were synonymized by re-examining type material and protologue. The ITS-based phylogenetic analysis placed its photobiont Trentepohlia sp. in a clade containing the unidentified Trentepohlia isolates TreFl63 (from Graphis sp., Brazil), TreFl149 (from Arthonia atra, UK), S16_Gp (from Graphis propinqua, Argentina), TreFl51 (from Diorygma pruinosum, Brazil), and TreFl61 (from Graphis sp., Brazil), suggesting this clade to be entirely lichenized.
-
Key words:
- Lichenized fungi /
- Symbiosis /
- Synonymization /
- Polyphasic taxonomy