HTML
-
Endophytic microorganisms colonize in plant tissues in which they spend part or all their life cycle without causing disease symptoms in the host (Petrini 1991). Fungal endophytes may inhabit in different organs of the host including leaves, stems, bark, roots, fruits, flowers and seeds (Rodriguez et al. 2009). Generally, in this symbiotic relationship fungal endophytes receive shelter and nutrients from the host, while the host plant may benefit from an array of attributes which include protection against natural enemies such as pathogens and herbivores (Schardl et al. 2004, Singh et al. 2011), plant growth promotion (Hamayun et al. 2010) and increasing the resistance of plants to abiotic stresses such as salinity and heavy metal toxicity in soil (Khan et al. 2014). Some medicinal plants are known for harboring endophytic fungi, which are important sources of various bioactive secondary metabolites and enzymes valuable for the pharmaceutical industry (Zou et al. 2000, Strobel et al. 2004, Krishnamurthy et al. 2008).
Endophytic fungi are relatively unexplored producers of metabolites useful in pharmaceutical and agricultural industries. A single endophyte can produce several bioactive metabolites. As a result, the role of endophytes in the production of various natural products with greater bioactivity have received increased attention (Prabavathy & Valli 2012). Pectinases and cellulases, besides other enzymes, are the most important enzymes produced by endophytic fungi as a resistance mechanism against pathogenic invasion and to obtain nutrition from the host. These enzymes have various industrial applications, thus of major interest. Increasing efforts are being taken to characterize and identify endophytic fungi from medicinal plants. Therefore, the present work was designed to study the biodiversity of endophytic fungi in some wild medicinal plants from the New Valley Governorate, Egypt, and to evaluate their ability to produce extracellular pectinases and cellulases.
-
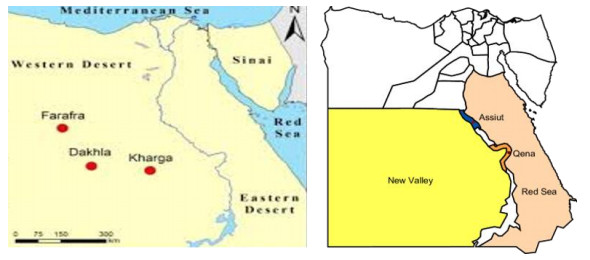
The New Valley Governorate is located at the Western Desert of Egypt. It encompasses 440, 098 km2, which is approximately 44% of the total area of Egypt and 66% of the area of Western Sahara. It is demarcated by the Governorates of Minya, Assiut, Sohag, Qena and Aswan from the east, by Libya and the Governorates of Matrouh and the Marine Oasis of the 6th of October City from the West and by Sudan from the South. The New Valley includes four large Oases namely El-Kharga (the sampling sites), El-Dakhla, El-Bahariya and El-Farafra, and the capital is El-Kharga (Fig. 1).
Sample collection and identification of plant species
-
Healthy and mature plant leaves and roots of eight wild medicinal plants were collected from El-Kharga Oasis, New Valley Governorate once during April 2018. Ten replicates from each of Alhagi graecorum, Anagallis arvensis, Calotropis procera, Chenopodium ambrosioides, Convolvulus arvensis, Moringa oleifera, Portulaca oleracea, and Ricinus communis plants were collected in sterile polyethylene bags and promptly brought to the laboratory for isolation of fungi. The plant species collected in the current investigation were identified according to morphological features and taxonomical characters at the Assiut University Herbarium, Department of Botany and Microbiology, Faculty of Science, Assiut University, Assiut, Egypt (Fig. 2).

Figure 2. Wild medicinal plant species collected from El-Kharga Oasis, the New Valley Governorate, Egypt, during April 2018.
Sample preparation and surface sterilization
-
Prior to surface sterilization, leaves and roots of each sample were thoroughly washed with tap water to remove the dust followed by distilled water. The samples were then cut into 5-cm segments. The samples were surface sterilized using the following sequence; 5% sodium hypochlorite for 3 min, 70% ethanol for 1 min, and washing with sterile distilled water 3 times each for 1 min. In aseptic conditions, both ends of each segment (1 cm) was cut off to produce a 3-cm segments (Al-Bedak et al. 2020).
Isolation of endophytic fungi
-
Segments of each sample were plated on Petri-dishes containing 1% glucose-Cz with the following composition (g/l): Glucose, 10; Na2NO3, 2; K2HPO4, 1; KCl, 0.5; MgSO4.7H2O, 0.5; FeSO4, 0.01; ZnSO4, 0.01; CuSO4, 0.005; Rose Bengal, 0.05; chloramphenicol, 0.25; agar, 15 and the final pH 7.3 (Ismail et al. 2017). The plates were incubated for 7-21 days at 25℃. Counts of CFUs of each fungal isolate were calculated per 25 segments in every sample. The obtained fungi were identified morphologically to the species level at the Assiut University Mycological Centre according to their macroscopic and microscopic characteristics. Pure cultures of the fungal strains were preserved for further investigations on PDA slants, as well as on cotton balls (Al-Bedak et al. 2019) at 4℃ in the culture collection of the Assiut University Mycological Centre.
Phenotypic identification of fungi
-
The obtained fungi in this study were identified morphologically to the species level at the Assiut University Mycological Centre according to their macroscopic and microscopic characteristics. The following references were used for the identification of fungal genera and species (purely morphologically, based on macroscopic and microscopic features): Booth (1971), Ellis (1976), Pitt (1979), Domsch et al. (2007), Moubasher (1993), de Hoog et al. (2000), Samson et al. (2004), Leslie & Summerell (2006), Simmons (2007) and Al-Bedak et al. (2020).
Screening of pectinase and endoglucanase production on solid medium
-
Production of pectinase and endoglucanase was detected on sucrose-free Czapek's agar medium amended with pectin (from citrus peel) and CMC as a sole carbon source, respectively. 50 µl of spore suspension from 7-day-old culture of each fungal strain was individually added to each 5-mm diameter well on the agar plate (Moubasher et al. 2016). The inoculated plates were incubated for 2 days at 30ºC. The clear zones formed around the wells were more visible when the plates were flooded with 0.25% (w/v) aqueous iodine solution. The diameters of the clear zones were measured (in mm) against the brown color of the test medium indicating enzyme production.
Production of pectinases and cellulases under submerged fermentation
-
All positive fungal strains were grown, individually in 250-ml Erlenmeyer conical flasks each containing 50 ml sucrose-free Czapek's broth medium supplemented with 1% pectin or 1% CMC as sole carbon source. The flasks were then inoculated individually with 1 ml spore suspension containing 1 x 107 spore/ml of 7-day-old culture of the tested strains. The inoculated flasks were then incubated at 30ºC in shaking condition of 150 rpm for 7 days.
Enzyme extraction
-
After incubation period, the flasks contents were individually filtered through filter papers (Whatman No. 1) and the filtrate was then centrifuged at 10000 xg for 10 min at 4ºC. The clear supernatants were used as a source for CMCase or pectinase enzyme.
Pectinase assay
-
The enzyme production was determined by mixing 0.9 ml of 1% pectin (prepared in 50 mM Na-citrate buffer, pH 5.0) with 0.1 ml of filtered crude enzyme, and the mixture was incubated at 50℃ for 15 min in a water bath (Bailey et al. 1992). The reaction was stopped by the addition of 2 ml of 3, 5-dinitrosalicylic acid (DNS) and the contents were boiled in water bath for 10 min (Miller 1959). After cooling, absorbance was measured at 540 nm using Cary 60 UV-Vis spectrophotometer. The amount of reducing sugar liberated was quantified using calibration curve of glucose. One unit of pectinase is defined as the amount of enzyme that liberates 1 µmol of glucose equivalents per minute under the standard assay conditions.
Cellulases (CMCase and avicellase) assay
-
The cellulases activity was determined by mixing 0.9 ml of 1% CMC or 1% avicel (prepared in 50 mM Na-citrate buffer, pH 5.0) with 0.1 ml of filtered crude enzyme, and the mixture was incubated at 50℃ for 15 min in a water bath (Bailey et al. 1992). The reaction was stopped by the addition of 2 ml of 3, 5-dinitrosalicylic acid (DNS) and the contents were boiled in water bath for 10 min (Miller 1959). After cooling, absorbance of the developed color was measured at 540 nm using Cary 60 UV-Vis spectrophotometer. The amount of reducing sugar liberated was quantified using calibration curve of glucose. One unit of CMCase or avicellase is defined as the amount of enzyme that liberates 1 µmol of glucose equivalents per minute under the standard assay conditions. Glucose concentration was calculated using the calibration curve.
$ {\bf { Glucose\; concentration }}=\frac{\text { Absorbance }}{\text { slope }(=1.0472)}\; \mathrm{mg} / \mathrm{ml}\;(=\mathrm{g} / \mathrm{L}) \\ {\bf { Enzyme \;concentration }}=\frac{\text { Glucose concentration }(\mathrm{g} / \mathrm{L})}{0.00018}\; \mathrm{IU} / \mathrm{L} $ The enzyme activity (pectinase or CMCase or avicellase) was calculated according to the following formula (Moubasher et al. 2016)
$ {\bf{Enzyme activity}}{\rm{ }} = {\rm{ }}{\bf{Absorbance x DF x}}\left({\frac{{\bf{1}}}{\mathit{\boldsymbol{x}}}} \right)\left({\frac{{\bf{1}}}{\mathit{\boldsymbol{y}}}} \right)\left({\frac{{\bf{1}}}{\mathit{\boldsymbol{t}}}} \right)\left({\frac{{\bf{1}}}{{{\rm{ }}\mathit{\boldsymbol{slope}}{\rm{ }}}}} \right) $ Where: DF = the dilution factor for enzyme, x = the volume of enzyme used, y = the volume of hydrolysate used for assay of reducing sugars, t = the time of hydrolysis, slope is determined from a standard curve
Sampling area
-
A total of 32 species related to 18 genera of endophytic mycobiota were recovered on 1% glucose-Cz at 25℃ from healthy and mature plant leaves and roots of eight wild medicinal plants, collected from El-Kharga Oasis, the New Valley Governorate. The high incidence in genera were recorded in Alternaria, Aspergillus and Fusarium. Fusarium (represented by 2 species) was the most common and encountered total CFU constituting 37.0% of total fungi. It was recovered from 7 plants out of 8. F. oxysporum was the most prevalent species encountering 23.1% of total fungi, however it was recorded from 3 plants only, followed by F. solani giving rise to 13.9% of total fungi and it was the most frequent recovered from 6 plants. Alternaria (7 species) came next to Fusarium and it was comprised 24.0% of total fungi with A. alternata being the most common Alternaria species recorded from 4 plants and was comprised 10.33% of total fungi followed by A. tenuissima (from 4 plants) comprising 7.42% of total fungi. Aspergillus (5 species in addition to 2 unknown species) was the runner of Alternaria comprising 17.5% of total fungi. It was the most frequent genus isolated from all the studied plants. The most prevalent Aspergillus species were A. terreus followed by A. flavus constituting 7.42% and 5.84% of total fungi respectively (Table 1).
Table 1. CFUs (calculated to the total CFUs of each fungus per 25 segments of leaves (L) or roots (R) of each plant sample), Gross total CFUs and % gross total CFUs of fungi isolated from 8 wild medicinal plants collected from El-Kharga Oasis, New Valley Governorate on 1 % glucose-Cz at 25℃ during April 2018.
Fungal genera & species Plant species Gross total Alhagi graecorum Convolvulus arvensis Chenopodium ambrosioides Calotropis procera Ricinus communis Angallis arvensis Moringa oleifera Portulaca oleracea L R L R L R L R L R L R L R L R CFU %CFU Acremonium 4 4 0.89 A. rutilum 1 1 0.22 A. sclerotigenum 3 3 0.67 Alternaria 1 34 36 1 20 15 107 24 A. alternata 1 21 20 4 46 10.33 A. brassicicola 1 1 0.22 A. chlamydospora 8 8 1.79 A. citri 1 7 8 1.79 A. citri macularis 4 4 0.89 A. longipes 3 3 1 7 1.57 A. tenuissima 10 8 9 6 33 7.42 Aspergillus 1 10 3 2 1 1 20 10 21 2 2 2 1 2 78 17.5 A. creber 2 2 0.44 A. flavus 1 1 1 19 2 1 1 26 5.84 A. fumigatus 1 1 2 0.44 A. keveii 1 1 0.22 A. parasiticus 1 8 1 1 1 12 2.70 A. terreus 1 9 1 20 1 1 33 7.42 A. tubingensis 2 2 0.44 Beauveria bassiana 1 1 0.22 Chaetomium senegalense 1 1 0.22 Cladosporium exile 1 1 0.22 Clonostachys solani 1 1 0.22 Curvularia spicifera 19 1 20 4.49 Fusarium 7 33 37 14 12 14 22 6 20 165 37 F. oxysporum 6 33 36 22 6 103 23.1 F. solani 1 1 14 12 14 20 62 13.9 Macrophomina phaseolina 1 4 1 1 7 1.57 Penicillium olsonii 21 21 4.72 Pseudoallescheria boydii 2 2 0.44 Rhizoctonia solani 1 2 3 0.66 Rhizopus microspores 1 1 0.22 Sarocladium kiliense 2 2 0.44 Scopulariopsis fimicola 1 1 0.22 Stemphylium botryosum 6 1 6 13 2.92 Verticillium fungicola 2 2 0.44 Yeast spp. 1 1 1 1 1 1 8 1 15 3.37 CFUs 1 37 43 44 36 20 62 23 45 21 52 9 18 0 10 24 445 100 No. of genera 1 4 5 6 3 4 3 3 5 6 6 3 3 0 2 4 No. of species 1 6 7 8 5 4 7 4 7 6 9 3 6 0 2 5 Total CFUs 38 87 56 85 66 61 18 34 445 Total genera (18) 4 9 6 5 10 7 3 5 Total species (32) 6 14 9 11 12 10 6 6 Aspergillus parasiticus, Macrophomina phaseolina were found in 4 plant species, A. longipes and Stemphylium botryosum in 3 plants, A. citri, A. fumigatus, and C. spicifera in 2 plants while Acremonium rutilum, Beauveria bassiana, Chaetomium senegalense, Cladosporium exile, Clonostachys rosea, C. solani, Pseudoallescheria boydii, Rhizoctonia solani, Rhizopus microsporus, Scopulariopsis fimicola, Stemphylium botryosum and Verticillium fungicola were recorded each in one plant species. Convolvulus arvensis was the richest plant with endophytes containing 14 species belonged to 8 genera and recording the highest CFUs of 79 per 25 segments over the remaining plant species, while Moringa oleifera was the poorest in endophytes with 5 species belonging to 2 genera and the lowest CFUs of 13 per 25 segments. It is worth mentioning that Beauveria bassiana; the known entomopathogenic fungus was recorded for the first time from leaves of Portulaca oleracea as an endophyte (Table 1).
Preliminary screening of endophytic fungi for pectinases and cellulases production
-
One-hundred and twenty fungal isolates representing 31 species related to 17 genera of endophytic fungi were screened for their abilities to produce pectinase and endoglucanase on sucrose free-Cz supplemented with 1% pectin or 1% CMC as a sole carbon source, respectively. Ninety-four isolates could produce pectinase enzyme, of which 18 were high producers, 25 moderate and 51 low. 66 isolates could produce cellulase, of which 13 were high producers, 16 moderate and 37 low (Appendix 1).
Table Appendix 1. Preliminary screening of pectinases and cellulases production by endophytic fungi recovered from leaves and roots of eight wild medicinal plants collected from El-Kharga Oasis, the New Valley Governorate, Egypt, during April 2018.
Fungal species Number of isolates tested Preliminary screening Pectinases Cellulases Positive L M H Positive L M H Acremonium 2 2 1 1 2 1 1 Acremonium rutilum 1 1 1 1 1 Acremonium sclerotigenum 1 1 1 1 1 Alternaria 26 20 14 6 6 4 2 A. alternata 8 7 5 2 2 2 A. brassicicola 1 1 1 A. chlamydospora 1 1 1 1 1 A. citri 3 2 2 A. citri macularis 2 2 2 A. longipes 3 3 1 2 1 1 A. tenuissima 8 6 6 Aspergillus 35 27 11 9 7 21 6 7 8 A. flavus 9 7 3 2 2 4 1 2 1 A. fumigatus 2 1 1 2 1 1 A. parasiticus 6 3 2 1 2 2 A. terreus 15 14 5 5 4 12 4 2 6 A. tubingensis 1 Aspergillus AY-1 1 1 1 1 1 Aspergillus AY-2 1 1 1 Beauveria bassiana 1 1 1 Chaetomium senegalense 1 1 1 Cladosporium exile 1 1 1 1 1 Clonostachys solani 1 1 1 1 1 Curvularia spicifera 2 1 1 2 2 Fusarium 23 20 14 3 3 16 13 1 2 F. oxysporum 13 11 8 2 1 11 10 1 F. solani 10 9 6 1 2 5 3 1 1 Macrophomina phaseolina 7 3 3 3 2 1 Penicillium olsonii 1 1 1 1 1 Pseudo allescheria boydii 1 1 1 1 1 Rhizopus microsporus 1 1 1 1 1 Sarocladium kiliense 1 1 1 1 1 Scopulariopsis fimicola 1 Stemphylium botryosum 5 5 3 2 3 3 Verticillium fungicola 1 1 1 1 1 Yeast spp. 10 7 3 1 3 6 4 2 Total isolates 120 94 51 25 18 66 37 16 13 No. of genera 17 16 10 8 8 14 9 6 5 No. of species 31 28 17 15 9 23 14 9 8 Note: H = high producers: ≥ 20 mm, M = moderate: 11-19 mm, L = < 11 mm Submerged production of pectinases and cellulases (CMCase and avicellase)
-
The quantitative assay of pectinase, CMCase and avicellase for high-producing isolates were performed in submerged fermentation using sucrose-free Cz broth medium amended with 1% pectin or CMC or avicel as the sole carbon source. Of these, 17 isolates could produce pectinase enzyme with a relative activity ranged from 147.84 IU/ml/min to 225.43 IU/ml/min while 14 could produce CMCase (1.84 IU/ml/min – 22.0 IU/ml/min) and avicellase (26.0 IU/ml/min – 47.87 IU/ml/min). Six isolates were found to have the abilities to produce the three enzymes, of which Aspergillus was the superior with the potent strains were A. terreus AUMC 14278 for pectinase activity giving 225.43 IU/ml/min and A. terreus AUMC 14287 for CMCase producing 22.0 IU/ml/min and avicellase recording 47.868 IU/ml/min (Tables 2-4).
Table 2. Pectinase production and activity of some endophytic fungi.
Fungal species AUMC no. Pectinase Glucose g/l Production IU/ml Activity IU/ml/min Aspergillus flavus 14274 19.27 107.063 147.8 A. flavus 14289 27.82 154.555 213.4 A. fumigatus 14283 27.44 152.438 210.5 A. terreus* 14278 29.4 163.244 225.4 A. terreus 14287 26.34 146.326 202.0 A. terreus 14293 26.0 144.814 200.0 A. terreus 14279 24.14 134.103 185.2 Cladosporium exile 14294 23.91 132.846 183.4 Curvularia spicifera 14276 23.6 131.016 180.9 C. spicifera 14273 29.0 161.255 222.7 Fusarium solani 14277 23.3 129.472 178.8 F. solani 14292 21.6 119.954 165.6 Macrophomina phaseolina 14272 23.253 129.185 178.4 M. phaseolina 14275 23.0 127.854 176.5 Penicillium olsonii 14295 23.73 131.843 182.0 Yeast sp. 14289 24.85 138.066 190.7 Yeast sp. 14281 22.9 127.275 175.8 * The highest producer showed in bold Table 3. Endoglucanase (CMCase) production and activity of some endophytic fungi.
Fungal species AUMC no. Endoglucanase (CMCase) Glucose g/l Production IU/ml Activity IU/ml/min Aspergillus flavus 14274 0.41 2.282 3.15 A. fumigatus 14283 0.3 1.684 2.32 A. terreus 14278 1.9 10.623 14.7 A. terreus* 14287 2.874 15.966 22.0 A. terreus 14280 2.68 14.895 20.6 A. terreus 14282 1.47 8.164 11.3 A. terreus 14284 1.9 10.579 14.6 A. terreus 14285 1.5 8.328 11.5 A. terreus 14288 1.0 5.575 7.7 Clonostachys rosea 14291 0.24 1.332 1.84 Curvularia spicifera 14276 1.3 7.167 9.9 C. spicifera 14273 1.39 7.711 10.65 Fusarium oxysporum 14290 0.757 4.206 5.8 F. solani 14286 0.7 3.910 5.4 * The highest producer showed in bold Table 4. Avicellase production and activity of some endophytic fungi.
Fungal species AUMC no. Avicellase Glucose g/l Production IU/ml Activity IU/ml/min Aspergillus flavus 14274 4.213 23.405.5 35.55 A. fumigatus 14283 4.232 23.512 35.71 A. terreus 14278 4.049 22.496 34.17 A. terreus* 14287 5.672 31.51 47.87 A. terreus 14280 4.418 24.51 37.23 A. terreus 14282 3.706 20.588 31.3 A. terreus 14284 3.671 20.395 30.98 A. terreus 14285 3.876 21.53 32.71 A. terreus 14288 4.907 27.258 41.41 Clonostachys rosea 14291 3.466 19.258 29.25 Curvularia spicifera 14276 3.592 19.958 30.32 C. spicifera 14273 3.782 21.008 31.91 Fusarium oxysporum 14290 3.085 17.139 26.0 F. solani 14286 3.709 20605 31.3 * The highest producer showed in bold
Biodiversity of endophytic fungi
-
In the current study, endophytic mycobiota in healthy and mature leaves and roots of eight wild medicinal plants were isolated on 1% glucose-Cz at 25℃ from sample collected once in April 2018. This study is considered as the first in the New Valley Governorate, Egypt for evaluation of endophytic fungi from these medicinal plants. There is a growing body of literatures that recognize the importance of endophytic fungi across a number of disciplines in recent years as biological sources of a wide range of valuable compounds including plant growth regulatory, antibacterial, antifungal, antiviral, insecticidal substances to enhance the growth and competitiveness of the host in nature (Anwar et al. 2007, Kaur & Kalia 2012, Khairnar et al. 2012, Al-Snafi 2015, Muhammad et al. 2015, Syed et al. 2016, Khan Marwat et al. 2017).
The current results revealed that endophytic fungal assemblages were obtained from all plant species examined and some plants were occupying by the same fungal genera and species, indicating that endophytic fungi can be the same in plants belonging to different families. Altogether, 32 species related to 18 genera were recovered from the leaves and roots of all tested plants.
The high occurrence genera where described by Fusarium, Aspergillus and Alternaria. Fusarium was the most widespread genus retrieved from 7 plants. F. oxysporum is the most dominant led by F. solani. Such latest observations have, to some degree, been compatible with the reports of Raviraja (2005) who researched endophytic fungi in five Brazilian medicinal plants and found that Aspergillus and Penicillium were isolated at high frequencies, however, Fusarium oxysporum was reported at low levels from leaves of two plants tested. Previous studies on plants of the same size as ours have previously been conducted with the genera Fusarium, Aspergillus, Nigrospora, Stachybotrys, Rhizoctonia and Macrophomina from Moringa leaves (Carbungco et al. 2015). Almost similar results were obtained in other studies on Calotropis procera in Karachi (Khan et al. 2007) or in Saudi Arabia (Gherbawy & Gashgari 2014).
Endophytic fungi produce enzymes such as amylases, cellulase, lipases and proteases, as part of their mechanism to overcome the defense of the host against microbial invasion and to obtain nutrients for their development (Patil et al. 2015). In addition, these enzymes are essential for endophytic fungi to colonize in the plant tissue (Sunitha et al. 2013). The array of enzymes produced differs between fungi and often depends on the host and their ecological factors (Sunitha et al. 2013). In the current study, 120 fungal isolates were screened for their ability to produce pectinase and cellulase. The results obtained revealed that 78.0% of the total isolates tested could produce pectinase enzyme and 55.0% could produce cellulase enzyme.
The quantitative assay of the three enzymes for high-producers were performed in submerged fermentation using sucrose-free Cz broth. Aspergillus was superior in the production of the three enzymes with the potent strains were A. terreus AUMC 14287 for CMCase and avicellase, and A. terreus AUMC 14278 for pectinase. Almost similar results were reported by (Sunitha et al. 2013) who found that 62.0% and 32.0% of their tested endophytic isolates were positive for pectinase and cellulase respectively, however, their tested fungi were isolated from plants differ from ours. In another study of cellulase activity of fungi inhabiting salt marshes, 100% of the tested isolates showed cellulolytic activity (Gessner 1980), while 66.0 % of fungi isolated from Brucea javanica could produce cellulase enzyme (Choi et al. 2005). The main endophytic fungi work in literature involves screening for secondary metabolites of antimicrobial and antioxidant activity. Not many explored the possibility of endophytic fungi as industrially essential biotechnological reservoirs of enzymes.
Cellulases have been widely used in agricultural, biofuel, detergent, fermentation, food, paper pulp, and textile industries (Kuhad et al. 2011). Screening of the isolates for cellulase activity was attempted with a view of endophytes penetrating the plant tissue through the lignocellulosic wall with the help of the hydrolytic enzymes, cellulases being predominant among them (Carroll & Petrini 1983). In addition, it was reported that some endophytes might behave as latent saprophytes, and when the host dies, they use these enzymes for tissue degradation to obtain nutrients (De Aldana et al. 2013). Studies also estimated that microbial pectinase accounts for 25% of global food and industrial enzymes revenues and is increasingly growing in the market (Oumer 2017). In addition, enzymes are a well-established global industry that is expected to hit USD 6.3 billion in 2021 (Oumer & Abate 2018).
The current results revealed that 78.3% of total isolates could hydrolyze pectin in submerged fermentation, of which 77.14% were Aspergillus isolates, 76.9% Alternaria, and 86.95% Fusarium showed positive results. The present findings were in concurrence with those of Sunitha et al. (2013) who reported that 62% of their tested fungi were pectinase producers, and better than results obtained by Shubha & Srinivas (2017) who found that 30% of their tested fungi had the pectinolytic activity. However, Choi et al. (2005) have reported that pectinase production was absent in all the endophytic fungi of Brucea javanica.
Aspergillus species was superior in pectinase activity with A. terreus being the potent strain giving rise to 163.244 IU/ml which is more than the result of pectinase production (106.7 IU/ml) produced by Aspergillus sp. Gm (KC et al. 2020) and much more the outcome of pectinase production (1.524 IU/ml) stated by (Sopalun & Iamtham 2020) from endophytic fungi isolated from Thai Orchids.
The production of plant cell-wall digestive enzymes is now a focus of current research. Many such researches have been done into the production of cellulase and pectinase due to the huge number of application scenarios of these enzymes (Jalis et al. 2014, Edor et al. 2018, Ismail et al. 2018, Li et al. 2020, Xue et al. 2020).
-
The current research investigates the ecology of endophytic fungi in wild medicinal plants in the New Valley Governorate, Egypt and determines their ability to produce hydrolyzing enzymes. The study managed to retrieve a total of 120 fungal isolates from just eight plants, indicating their widespread distribution. The study also confirmed the ability of these fungal isolates to produce pectinase and cellulase. The potent strains of Aspergillus was the superior in enzymes production with A. terreus AUMC 14287 for CMCase and avicellase, and A. terreus AUMC 14278 for pectinase. The study further highlights the promising ability to produce extracellular enzymes by endophytic fungi, thus showing the importance of further analysis to resolve key issues in this area.
-
The authors have not declared any conflict of interests.
- Copyright: © 2021 by the author(s). This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
| MA Abdel-Sater, AMA Abdel-Latif, DA Abdel-Wahab, OA Al-Bedak. 2021. Endophytic mycobiota of wild medicinal plants from New Valley Governorate, Egypt and quantitative assessment of their cell wall degrading enzymes. Studies in Fungi 6(1):78−91 doi: 10.5943/sif/6/1/4 |