-
Chrysanthemum morifolium is an important ornamental flower and potted plant with large demand in the consumer market[1]. In order to address growing consumer needs, it's necessary to increase the quality and resistance of existing species to biological or abiotic stress[2]. Classical breeding is used to improve series of characteristics, but due to the cross barriers, the use of this technique is limited. Fortunately, there is another way to create variation by introducing specific features through genetic engineering[3,4]. Genetic transformation of chrysanthemums prevalent using leaf explants has been performed by many researchers[5,6]. This approach requires efficiency in complicated vitro bud differentiation and regeneration procedures, it's easy for the leaf discs to accept foreign genes, but the transformation efficiency rate is low being from 0.1% to 6.25%[7−9], and it often produces lots of chimeras and false positives[8]. In addition, it usually takes at least 4−6 months to obtain transgenic plants with leaf disc transformation if all goes well, but the time is often lengthier as there are usually frequent failures during the complicated transformation procedures[9,10]. Thus, this method is time consuming. Another approach is protoplast transformation with a high transformation efficiency and no chimeras, but the process of protoplast culture is difficult, the operation is complicated and the frequency of regeneration is low. Thus, the application of protoplast transformation is seldom currently used[11,12]. Nowadays, one method widely used to enhance Agrobacterium infection is vacuum filtration, which has gained success to produce bean[13], Arabidopsis[14], banana[15], coffee[16] and Monterey pine[17]. Vacuum infection is an effective adjunct to improve Agrobacterium-mediated genetic transformation efficiency[18] by increasing the infiltration of Agrobacterium cells into the plant tissue layer[19]. However, up to now, the application of vacuum infiltration infection in chrysanthemum transgenic system establishment hasn't been reported, as well as the stem segment.
In this study, a highly efficient and useful Agrobacterium-mediated transformation protocol via vacuum infiltration was developed in chrysanthemum 'Yuhualuoying' using stem internode as explants. This protocol not only provides a new way for the efficient transformation of chrysanthemum and analysis of gene function, but also provides an inventive idea for the transgenic research of species without mature transformation systems.
-
Following 3 d pre-culture, no significant changes have taken place, but it's obvious that the stem internode explants were fresh and lively (Fig. 1a). The first morphogenetic change was observed during decarboxylation, for about 7 d, during which, the axillary buds of stem segments began to differentiate (Fig. 1b). Untransformed buds on selected medium had turned yellow and died due to the selection of hygromycin. However, the part of success could be multiplied rapidly to form a mass of shoots. Regeneration takes place in the tissue around the original buds. The axillary buds of the stem segments continued to differentiate into clusters in the course of subcultures (Fig. 1c). Turning to the rooting test, the escapes were unable to root on medium containing a high concentration of hygromycin (9 mg/L), while the transgenic shoots grew normally (Fig. 1d). As time progressed, the root system was stronger (Fig. 1e). Finally, the transgenic plant could be obtained (Fig. 1f).

Figure 1.
The regeneration of stem internode explants. (a) The explants were cut from seedlings with undifferentiated axillary buds and without stem tips and stem leaves. The best age of the materials is 30 d. (b) The axillary buds of stem segments began to differentiate on the medium containing 6-BA and NAA. (c) Transformed buds rapidly formed a mass of shoots. (d) The transgenic shoots were unable to root in the rooting test. (e) Obvious root systems of the transformation. (f) The transgenic plants.
Regeneration in the presence of hygromycin
-
Internode explants from in vitro grown stem material were co-cultivated with EHA105 containing the binary vector pMDC32 (Fig. 2), and hygromycin was used for selection because of the hygromycin resistance sequences on the foreign vector. During the training process, some of the explants died because of breakouts of Agrobacterium or bacterium from the environment. As the incubation time increases, the unpolluted explants have undergone different changes. Some stopped to elongate and turned yellow and died within 3−4 weeks. Some differentiated into clusters successfully, for each site, there were at least three shoots. From 100 explants in five petri dishes, 284 shoots regenerated in initial selection medium from 83 different stem segments. If the regeneration efficiency is defined as the number of shoots divided by the number of explants, this was 284%, while the survival rate was 83%. During every 15 d of subculture, the lateral buds were separated and endured selection pressure alone, some small shoots turned yellow without root formation. Finally, rooting to test was taken to exclude escapes. After several rounds of screening, there were 107 regeneration shoots left. Though regeneration buds are formed, those that can't carry out normal primordia differentiation on rooting test medium are classed as non-transformants. The transformation efficiency of resistant seedlings was 37.7%.
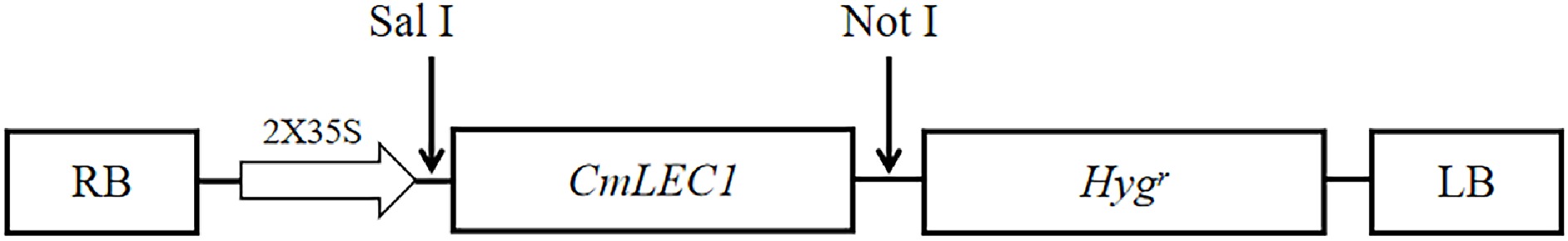
Figure 2.
Plant expression vector construction. Exogenous gene CmLEC1 was linked at the expression vector pMDC32, which contains the encoding sequence of hygromycin resistance. There is also a CaMV35S promoter, which can start the expression of CmLEC1.
Molecular analysis of the transformed plants by PCR
-
The transformants rooted in the pressure of 9 mg/L hygromycin were identified to test for the presence of the CmLEC1 transgene, The genomic DNA was isolated after 72 d from the infection to grow roots. As the result, among the 45 independent plantlets of regenerated plants, 19 resistant plantlets amplified a clear band, pointing to the 2,000 bp marker. The amplification length was the same as that of the positive control (Fig. 3). The PCR products were sequenced and proved to be consistent with the CmLEC1 sequence. It was preliminarily confirmed that the exogenous gene had been successfully transferred into chrysanthemum and 19 PCR positive single plants were obtained. The 19 transgenic lines were obtained by expansion and propagation of single plants and the positive seedling efficiency was 42.23%. This result indicated that transformation efficiency was reduced from 37.7% to 16%, if the influence of false positives events were removed.
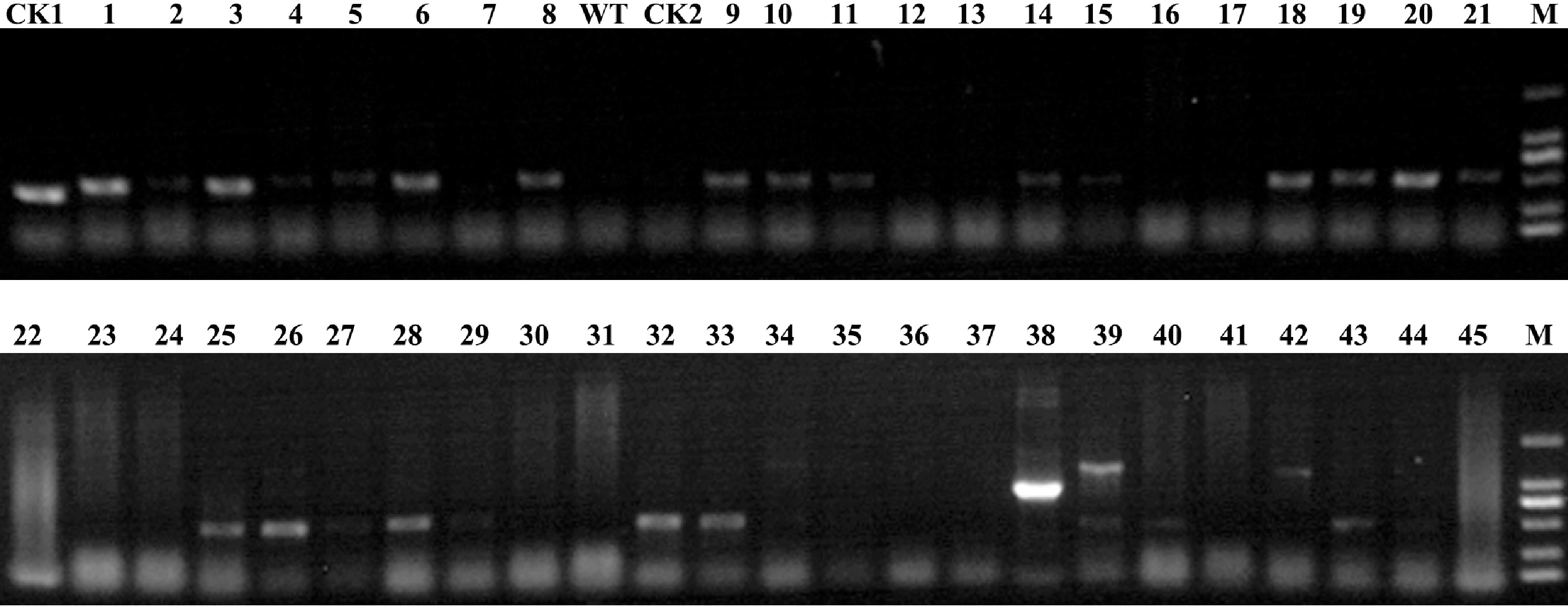
Figure 3.
Identification of transgenic plants at the DNA level. PCR analysis was carried out among the putative transgenic plants and control plants to confirm successful transformation. M: 2000 marker; 1−45: the putative transgenic plants to be identified; WT: 'Yuhualuoying' wild type plants; CK1: positive control; CK2: negative control.
Expression analysis of CmLEC1
-
Stable integration of the CmLEC1 can be confirmed by qRT-PCR analysis. The phenomena showed that, among the 13 positive plants which were identified, nine plants achieved a super expression of CmLEC1 more than two times higher, and five plants were five times higher than WT. What's more, the No.18 line had the highest expression, which was 21.86 times relative to WT, and followed by the No.11 plantlet, 15.10 times (Fig. 4). The results confirmed the presence of the CmLEC1 gene and proved the feasibility of this method, which can quickly obtain transgenic plants.
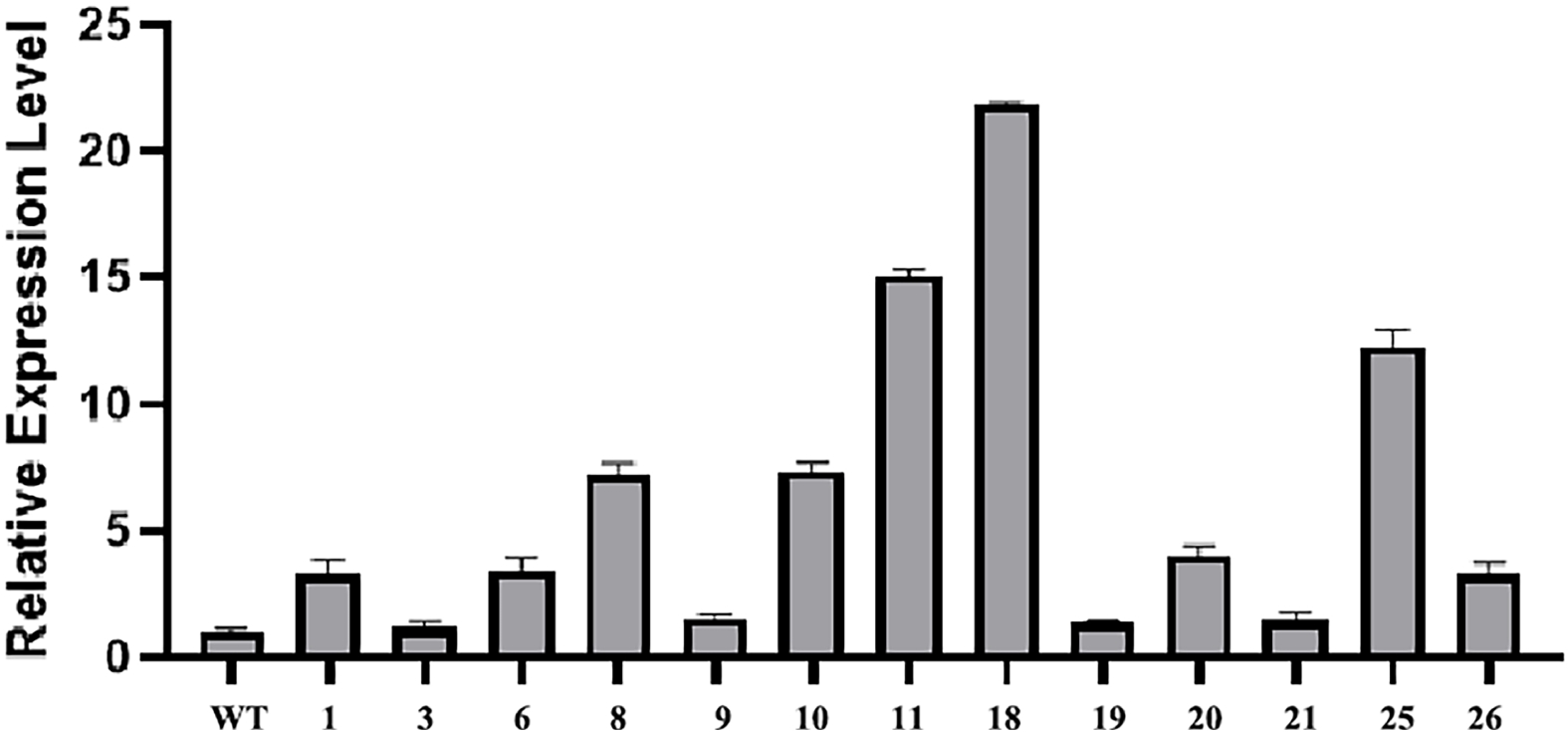
Figure 4.
Identification of positive transformants. Relative expression of CmLEC1 in chrysanthemum 'Yuhualuoying' was confirmed by qRT-PCR analysis.
DISCUSSION
-
Currently, genetically modified technology can be divided into physical, chemical and biological methods[20]. No matter what kind of method, they are all relying mainly on the dedifferentiation and re-differentiation of transformed cells in the tissue culture plant regeneration process to achieve the transfer of exogenous genes, and finally obtain transgenic plants. And there isn't currently a universal transgenic system[21].
The leaf disc method is one of the traditional transgenic methods, it's easy for the leaf discs to accept foreign genes, but the transformation efficiency rate is low. It's even less than 1% with some materials[7]. In our study, we used the chrysanthemum cultivar 'Yuhualuoying' as our material due to the continuity of the experiment. The previous physiology, cytology experiments and bioinformatics analysis were taken on the basis of this cultivar[2,22,23], and in order to determine the function of some important genes, transgenic research should be carried out. Under this circumstance, homologous transformation is the best choice to make the result convincing. But, there isn't a mature transgenic system for the chrysanthemum 'Yuhualuoying', and early exploration experiments undertaken showed that when the leaves were used as explants, the vitrification was serious and the transformation effect was not stable. After more than one year of study, we failed to obtain any transgenic plants with leaf disc transformation (data were not given). Thus, we needed to establish a stable system of regeneration and transformation from other perspectives.
As explant, the stem internode has a stronger ability to differentiate relative to the leaf[24], which is an advantageous feature for genetically modified implementation. So far, research has reported the use of stem segments as explants for transgenic studies on plants such as citrus, kumquat, potato and poplar[25−28], but the effect of infection and transformation is unsatisfactory due to the small incision area and other factors. In this study, we use the stem internode explants assisted with the vacuum method, to overcome the difficulties of infection.
Vacuum infiltration, as an accessorial method of in situ transformation, is simple, rapid and reliable. It opens up a new avenue for plant transgenic methods. However, in the past, researchers used a vacuum pump to improve the method of Agrobacterium infiltration[29], which was difficult to control. The intensity and time is difficult to determine, so that the material being dealt with may be easily destroyed and wasted. In this study, the transformation was carried out via manual vacuum infiltration treatments using common tools, which is visible and operable. We took 5−10 mins for vacuum infiltration until almost all stem sections sank to the bottom of the liquid. The stem of chrysanthemum 'Yuhualuoying' is soft and delicate, as other species have different degrees of lignification of stems, the processing time is flexible. Researchers are able to observe the status of the stem sections due to the manual handling. The highlight characteristics of this system is the combination of tissue location and vacuum infiltration. Vacuum infiltration can better help Agrobacterium enter the plants and the stem internode has a stronger ability to differentiate relative to the leaf.
According to the experimental results, the transformation efficiency of the methods provided in our article can be widely used in other species and cultivars with simplicity and ease of operation. What's more, the method provided in the article has higher efficiency. The regeneration efficiency was up to 284% as we used the stem internode containing undifferentiated axillary buds, which has a high differentiation ability, especially with the stimulating effect of plant hormones. At each site, there are many axillary buds occurring. But not all the shoots regenerated are positive ones, only the shoot tips that rooted in the presence of 9 mg/L hygromycin were classed as transformants, the transformation efficiency was 37.7%. If the escapes were removed, it was 16%, which was much higher than other methods. Generally, the transformation efficiency was about 4% on average using traditional transgenic methods[7], and the plant regeneration and gene transformation was affected by species, strains, pollution and many other factors, the transformation efficiency is even less than 1% in some cultivars[9]. Taking a cut chrysanthemum cultivar 'Jinba' for example, the transformation efficiency is only about 1% of its mature chrysanthemum system (experimental data from our laboratory). Agrobacterium-mediated transformation gives us a new idea for research on genetic function and improvement of biological traits. However, how to improve the efficiency of genetic transformation efficiency, shorten the time and reduce the workload are still the problems that we need to solve. In this study, we established an auxiliary method, i.e. vacuum infiltration, to more easily obtain transgenic plants compared to traditional methods.
-
Choosing the proper starting material is crucial to the entire regeneration[30]. In this study, Chrysanthemum cultivar 'Yuhualuoying' obtained from the Chrysanthemum Germplasm Resource Preserving Centre, Nanjing Agricultural University, China was chosen as the wild-type. The aseptic seedlings for genetic transformation were obtained depended on the space and technology provided by the Ornamental Horticulture Genetics and Breeding Laboratory of Nanjing Agricultural University. Young shoots were surface sterilized by initial immersion in 70% ethanol for 30 s, then in 30% H2O2 for 10 min, and finally being rinsed three times in sterile distilled water[31]. They were then incubated for 3 weeks on solidified Murashige and Skoog (MS) medium[32]. In order to obtain the best results, we took the tissue culture seedlings with the seedling age about 30 d as materials for subsequent experiments and operations (Fig. 5a).
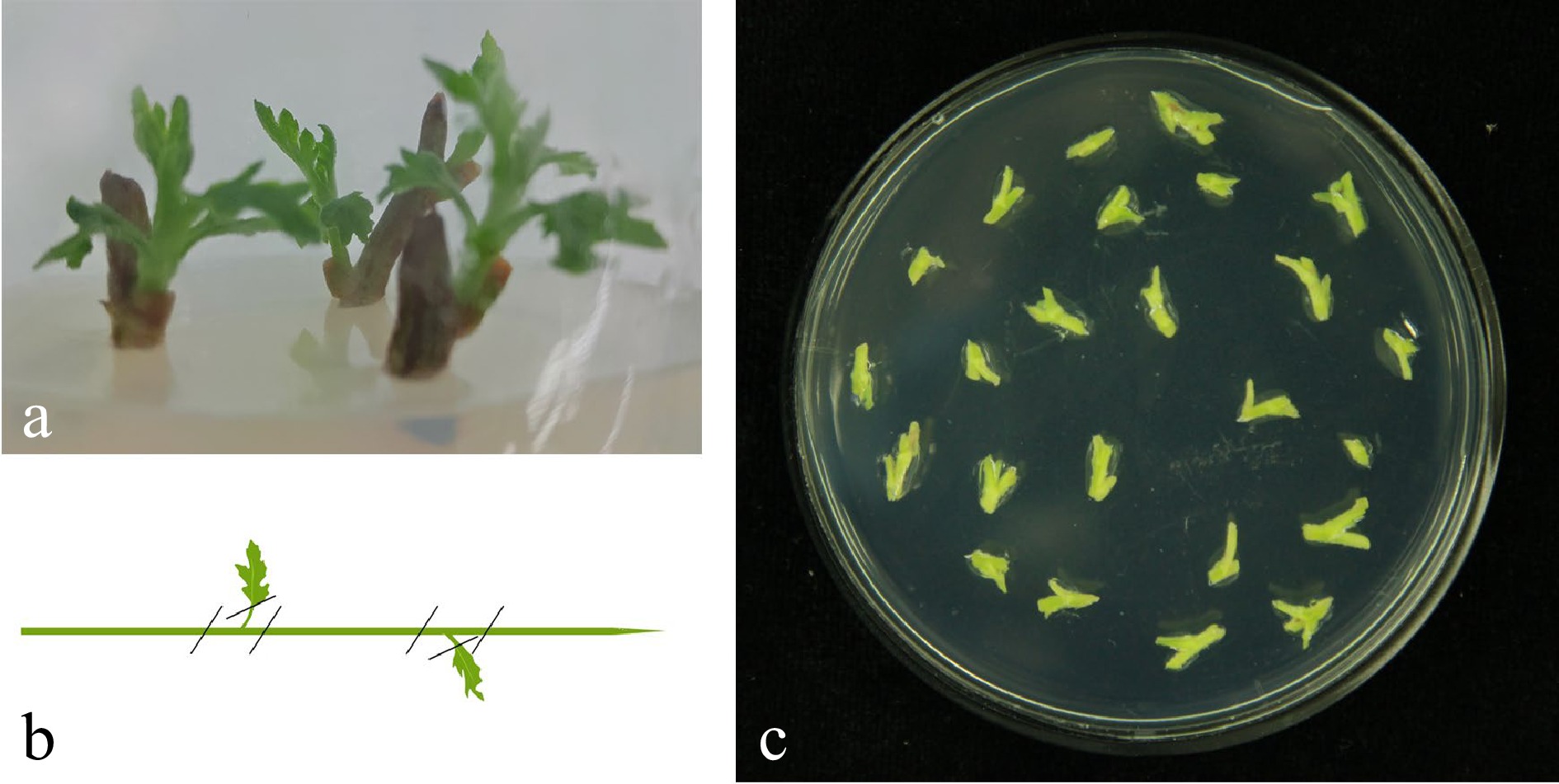
Figure 5.
Tissue culture seedlings from explants sterilized and stem material preparation. (a) Stem explants were taken from plants growing on land, young shoots were surface sterilized and tissue seedlings were cultured in a sterile environment. Note: steam internode explants with a seedling age of 30−40 d were used. (b) Remove the steam tips and steam leaves and cut the explants into 0.5 cm sections containing undifferentiated axillary buds. (c) Pre-culture the explants on MS basal medium containing 2.0 mg/L 6-BA and 0.1 mg/L NAA.
Recipes for modified MS medium in each step are listed in Table 1. MS basal medium was used to provide nutrients for culture seedlings, 6-benzylaminopurine (6-BA) and Napthalene acetic acid (NAA) are two hormones for dedifferentiation and differentiation of explants[33]. While Carbenicillin (Carb) was used for inhibiting the outbreak of Agrobacterium, and hygromycin (Hyg) was used as a selectable marker for transformation[10]. Throughout the whole cultivation process, the temperature was maintained at 25 °C. Except the co-cultivation stage, during which the materials were put under dark conditions at all times, there would be 16 h of illumination and 8 h in darkness set by hand. The plant hormones and chemical reagents used in this research were provided by Sigma, and the various enzymes were provided by Takara and Invitrogen.
Table 1. Several culture mediums used during the training process.
Medium name Code Medium composition Pre-culture and co-culture medium MS1 MS + 6-BA 2.0 mg/L + NAA 0.1 mg/L Decarboxylation medium MS2 MS + 6-BA 2.0 mg/L + NAA 0.1 mg/L + Carb 350 mg/L Select medium MS3 MS + 6-BA 2.0 mg/L + NAA 0.1 mg/L + Carb 350 mg/L Select medium MS4 MS + 6-BA 2.0 mg/L + NAA 0.1 mg/L + Carb 80 mg/L + Hyg 8 mg/L rooting medium MS5 MS + Hyg 9 mg/L MS: Murashige and Skoog basal medium (Potassium Nitrate 1,900 mg/L, Ammonium Nitrate 1,650 mg/L, Potassium Phosphate Monobasic 170 mg/L, Magnesium Sulfate 370 mg/L, Calcium Chloride 440 mg/L, Potassium Iodide 0.83 mg//L, Boric Acid 6.2 mg/L, Manganese Sulfate 22.3 mg/L, Zinc Sulfate 8.6 mg/L, Sodium Molybdate 0.25 mg/L, Cupric Sulfate 0.025 mg/L, Cobalt Chloride·6H2O 0.025 mg/L, Ferrous Sulfate 27.8 mg/L, Myo-Inositol 100 mg/L, Glycine 2 mg/L, Thiamine Hydrochloride 0.1 mg/L, Pyridoxine Hydrochloride 0.5 mg/L, Nicotinic Acid 0.5 mg/L, Sucrose 30 g/L, Sugar 6.5−7 g/L, PH = 5.8, adjusted with NaOH); 6-BA: 6-benzylaminopurine; Carb: Carbenicillin; NAA: Napthaleneacetic acid; Hyg: hygromycin. CmLEC1 gene isolation and vector construction
-
In order to achieve the CmLEC1 open reading frame (ORF), bioinformatics analysis was conducted according to the full length sequence of CL4474.Contig1 in 'Yuhualuoying' transcriptome database and NCBI[34]. The CmLEC1 fragments were obtained by polymerase chain reaction (PCR) with primers designed (Table 2) and RNA extracted from the embryo. Finally, we got the full length sequence of the gene, which is 1002 bp, and the ORF is 660 bp that can encode a protein of 219 amino acids. Specific primers were designed to import Sal I and Not I restriction sites, and link the CmLEC1 to the vector pENTR1A (Table 2). The amplicons were gelling purified and linearized by endonuclease PvuI. Then LR recombination reaction was carried out to subclone the amplicons into pMDC32 which contains the CaMV35S promoter and the nos terminator. The schematic diagram of the vector is shown in Fig. 2. Constructs were transformed into Agrobacterium tumefaciens strain EHA105 using a freeze–thaw method[35].
Table 2. Primers used in CmLEC1gene isolation and vector construction.
Primer Sequence (5'-3') CmLEC1 forward ATGGGTTACAATTGTGATTACTGTGG CmLEC1 reverse TCAGAAACTTGTTGCTTCATTCATGG CmLEC1-SalI forward TTCAGTCGACATGGGTTACAATTGTGATTA CmLEC1-NotI reverse GAGTGCGGCCGCGAGAAACTTGTTGCTTCATTCA Manual vacuum infiltration treatments
-
In this study, we took the healthy and thriving tissue culture seedlings with a seedling age about 30−40 d, removed the steam tips and steam leaves from the base of the petiole, and cut the explants into 0.5 cm sections containing undifferentiated axillary buds (Fig. 5b & c). MS basal medium containing 2.0 mg/L 6-BA and 0.1 mg/L NAA was used for pre-culturing the stem internode. One hundred cut stem segments were put on five petri dishes. This operation was repeated three times. Agrobacterium infection solution was used to introduce the CmLEC1 gene. Agrobacterium was activated and cultured overnight with YEB medium (Yeast extract and peptone) containing 50 mg/L kanamycin and 50 mg/L rifampin on an orbital shaker at 250 rpm at 28 °C (Fig. 6a). When OD600 reached 5.0−6.0, select bacteria by centrifugation at 4,000 rpms for 15−20 mins (Fig. 6b). Bacteria were resuspended in liquid MS medium to OD600 1.0 (Fig. 6c). After 3 d of pre-culture, the stem segments were picked from the solid medium and placed in a syringe (30 mL in volume, Fig. 6d). Then, approximately 8−10 mL of the infection solution was drawn in the syringe so that the stem section could be completely immersed in the infection liquid (Fig. 6e). We removed the needle, drained the gas in the syringe, and used the left thumb to block the front opening of the syringe, or burned the orifice impermeable with an alcohol lamp, then gently pinched and sealed (Fig. 6f). Then pulled the syringe plunger and shaked to release the interstitial air, which infiltrated the fluid into the organization simultaneously due to the negative pressure, this was carried out 5−8 times, for ten seconds each time. We used a 10mL tube as an aid to ease operation and hold the syringe, keep the negative pressure state, shake the syringe from time to time. This step was the most important for determining the effect of infection and transformation and 5−10 mins was necessary to ensure almost all steam sections sink into the bottom of the liquid (Fig. 6g). Then filtered off the infection liquid, put the infected stem segments on clean, sterilized filters (Fig. 6h), dried with sterile wind and replaced on new medium.

Figure 6.
Agrobacterium infection solution preparation and manual vacuum infiltration treatment. (a) Agrobacterium was cultured overnight with YEB medium. (b) Bacteria were selected by centrifugation. (c) Bacteria were resuspended in liquid MS medium. (d) Stem segments were picked and placed in a 30 mL syringe. (e) Infection liquid was drawn in the syringe. (f) Through a series of simple operations to create a vacuum environment. (g) Pull the syringe plunger and shake to release the interstitial air, keeping the negative pressure state for 5−10 min, until all steam sections sink to the bottom of the liquid. (h) Dry the infected stem and replace on new medium.
Culture process
-
The stem internode explants treated as above were pre-cultured on MS1 (Table 1) medium, containing 2.0 mg/L 6-BA and 0.1 mg/L NAA for 3 d. We then introduced the CmLEC1 by vacuum infiltration manually by the methods provided in this article, dried the infected stem segments and replaced on new MS1 medium, and co-cultivated with Agrobacterium containing the recombinant vector pMDC32-CmLEC1 for 2 or 3 d under dark conditions. When the edge of the explants showed spot-shaped Agrobacterium colonies, they were transferred to the Decarboxylation culture (MS2), in which Carbenicillin was used to inhibit the outbreak of A. tumefaciens (Table 1, 7 d or more is necessary). The infected stem segments were cultured and selected under the stress of hygromycin on MS3 and MS4 medium (Table 1)[36], and each 15 d was subcultured once. In each subculture, the small lateral buds should be separated from the mother and cultured separately, in order to screen continuously and reduce the production of transgenic chimerism. In the screening process, the concentration of the carboxybenzyl penicillin was reduced in sequence according to the gradient. The explants could be cultured on MS4 medium until rooting. If redisdual Agrobacterium was observed this should be remedied in time. The explants could be transferred to the rooting medium MS5 (Table 1) after a month without the outbreak of A. tumefaciens, with the aim of excluding false positives. Rooted plantlets were transplanted into sterile potting mixture and grown in a greenhouse.
Identification of transgenes
-
Transgenic plants were identified by confirming the certain presence of the CmLEC1 gene[25]. Among all the regenerated seedlings obtained by Agrobacterium infection and control plants (WT), genomic DNA was isolated. Some of the putative transformants were analysed by PCR amplification using a set of specific primers (Table 3), designed by the vector and gene[37]. Plasmid DNA was used as positive control (CK1), and clear water as negative control (CK2) to exclude the impact of environmental factors. PCR products were separated by agarose gel electrophoresis and visualized by ethidium bromide staining to confirm integration of CmLEC1 in possible transformants[38].
Table 3. Specific primers designed for identification.
Primer Sequence (5'-3') 35S GACGCACAATCCCACTATCC CmLEC1-TESTreverse ACATTCTTGGATGGTTTCTTTT CmLEC1-RT Forward GGCCATGAGCAAACTAGGGT CmLEC1-RT Reverse ACGTTCTCCGCCATCAAACT CmEF1α Forward TTTTGGTATCTGGTCCTGGAG CmEF1α Reverse CCATTCAAGCGACAGACTCA Transcription of transgenes
-
Stable expression of the chrysanthemum CmLEC1 in 'Yuhualuoying' was confirmed by qRT-PCR analysis. The relative expression analysis of CmLEC1 was carried out with wild-type chrysanthemum and transgenic chrysanthemum which was the same age and the same part, and there are three replications for each sample. Total RNA was extracted from with TRIzol reagent (TaKaRa, Tokyo, Japan) following the manufacturer's protocol. Single-stranded cDNA was obtained using an M-MLV Reverse Transcription Kit (TaKaRa), and qRT-PCR was implemented on a LightCycler 480 Real-Time PCR System. The gene-specific primers of CmLEC1 were designed (Table 3) by Primer 5[39]. And EF1α (Elongation Factor 1a) was used as a reference gene, which is stably expressed in chrysanthemum (sequences given in Table 3)[40,41]. The qRT-PCR was performed using a 20 μL reaction system. In addition, relative expression levels were calculated by the 2−ΔΔCᴛ method[42].
-
In summary, an efficient Agrobacterium-mediated transformation protocol via vacuum infiltration of chrysanthemum stem internode has been successfully established in the present study. This method not only significantly improves chrysanthemum transformation efficiency, but also dramatically shortens the time to obtain the transgenic positive seedling. Thus, the results of this study will be valuable and useful in gene function and molecular breeding of chrysanthemum, and other plants, in the future.
This study was supported by the National Natural Science Foundation of China (31672182, 31171983), the Priority Academic Program Development of Jiangsu Higher Education Institutions, and the Programs for New Century Excellent Talents in Universities, Ministry of Education of China (NCET-11-0669).
-
The authors declare that they have no conflict of interest.
- Copyright: © 2022 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Xu S, Wu Z, Zhao J, Zhang F, Zhang X, et al. 2022. High-efficiency Agrobacterium-mediated transformation of chrysanthemum via vacuum infiltration of internode. Ornamental Plant Research 2:1 doi: 10.48130/OPR-2022-0001
High-efficiency Agrobacterium-mediated transformation of chrysanthemum via vacuum infiltration of internode
- Received: 08 November 2021
- Accepted: 20 December 2021
- Published online: 18 January 2022
Abstract: Leaf disc transformation is one of the traditional methods that are now widely used in chrysanthemum with highly economical and ornamental value in world flower production, but it depends on plant genotypes and is time consuming and complicated. In addition, the transformation success rate of this method is low, generally ranging from 0.1% to 6.25%. Therefore, a highly efficient transformation system is needed. In this study, we are the first to establish a high-efficient chrysanthemum Agrobacterium-mediated transformation system via vacuum infiltration. Chrysanthemum stem internode explants were used as research material and CmLEC1 was used as a reporter gene. After approximately 3 months of culture and selection, the positive transgenic plants were obtained. Additionally, the positive probability was about 42%. The transformation efficiency was up to 37.7%, and if the escapes were removed, it was 16%. Furthermore, stable expression of CmLEC1 in transgenic 'Yuhualuoying' was confirmed by qRT-PCR analysis. These results suggest that this genetic transformation system via vacuum infiltration of chrysanthemum stem internode is highly efficient and convenient, and much better than traditional leaf disc transformation, and it will play an important role in chrysanthemum transformation and functional genetics research.
-
Key words:
- Agrobacterium /
- Transformation /
- Method /
- Vacuum /
- Chrysanthemum












