-
Cladosporium represents a diverse genus of Ascomycota. It possesses a distinctive conidiophore called coronate[1,2]. This fungus includes complex and large groups of species of about 772 names, and this number is likely to increase due to continued isolation from various substrate sources[3,4]. Cladosporium taxonomy has recently focused on phylogenetic relationships, and many studies have helped explain the taxonomy of this genus[5,6]. It is isolated from different substrates such as plants, water, air, food, soil, and clinical samples. It causes allergenic, opportunistic phaeohyphomycosis, and subcutaneous infections. Therefore many species are also isolated from the indoor environment. This shows the high adaptive ability of the fungus to revert to its physiology so that it can live as a saprophyte, pathogen, endophyte, and in extreme habitats[7−9]. Cladosporium has small conidia that easily spread over long distances, and it represents the most components isolated from air[10]. Cladosporium spp. found in conidial stage anamorph and the sexual stage teleomorph is rare, recently molecular studies demonstrate classification of sexual stage Davidiella[11]. Tibpromma et al.[12] isolated Cladosporium endophyticum in Thailand from Pandanus sp. as an endophytic fungus and described it as a new taxon. During a survey of airborne fungi in Basrah, Iraq, Cladosporium endophyticum was isolated.
This study aimed to identify C. endophyticum as the first record in Iraq and study the physiological factors that affect fungal radial growth.
-
Air specimens are collected using a petri dish of Potato Dextrose Agar. The Cladosporium isolates were purified and moved to a new PDA medium. Slide culture was prepared for observation and measurements of the microscopic feature.
Molecular study
-
The Fungus Genomic DNA Mini-Preps Kit extracted DNA from fungal colonies grown on potato dextrose agar (Biobasic, Canada). The ITS area was replicated utilizing primer ITS1/ITS4 and 50 µl of total PCR amplification volume[13]. Macrogen Company Korea carried out the sequencing.
The sequence data were processed using the program (DNASTAR, Madison, WI, USA) Version 7.1 and phylogenetic tree drawn according to the neighbor-joining method and visualized using the BLAST program[14−16].
Growth of fungi under various conditions of temperature, pH, medium, carbon, and nitrogen
-
A PDA medium was made to determine at how temperature affects the growth of fungal radial growth. A pure culture of C. endophyticum was cut into a 5 mm diameter disc and placed in the center of petri dishes. Then, the plates were placed in an incubator at 5, 10, 15, 20, 25, 30, 35, 40 °C, with three copies for each temperature, and the plates were kept there for two weeks. After 14 d, the size of the colonies were measured[17].
The influence of pH on fungal radial development was studied in PDA medium produced with varied pH values (4, 5, 6, 7, 8). A 5 mm-diameter disc was cut from a pure C. endophyticum culture and positioned on the middle of the petri plate. A 14-d dark incubation at 25 °C in triplicate with three dishes per pH followed. It was determined that the colony's diameter had grown after incubation[ 17,18].
Four solid media were produced to evaluate the influence of various media on fungal radial development. Malt extract agar (MEA), standard potato dextrose agar (PDA), synthetic nutrient-deficient agar (SNA) (glucose 0.2 g, sucrose 0.2 g, potassium dihydrogen phosphate 1 g, potassium nitrate 1 g, magnesium sulfate anhydrous 0.25 g, potassium chloride 0.5 g, agar 14 g, distilled water 1,000 ml), oatmeal agar (OA) (rolled oats 30 g, agar 15 g, distilled water 1,000 ml). The colony diameter was measured after a 5 mm diameter disc was cut from pure culture and placed in the middle of petri dishes and cultured in the dark for 14 d. There were three replicates for each medium.
To investigate the effect of carbon sources on fungal radial development, the various carbon sources were first introduced to a base medium (30 g·l−1): sucrose, glucose, sorbitol, xylose, and manitol (NaNO3 2.0 g, MgSO4·7H2O 0.5 g, K2HPO3 1.0 g, KCL 0.5 g, FeSO4 0.01 g, and agar 20 g purified water, 1,000 ml). At 25 °C, the petri dishes are incubated with a fungal disc diameter of 5 mm. After 14 d of incubation, the diameter of the colonies was measured[19].
Studying the effect of nitrogen sources on fungal development was carried out using czapeks medium (sucrose 30.0 g, MgSO4·7H2O 0.5 g, KCL 0.01 g, FeSO4 0.01 g, KH2HPO3 1.0 g, agar 20 g, and distilled water 1000 ml) as the foundation medium. Ammonium nitrate, ammonium sulfate, urea, ammonium phosphate, potassium nitrate, and ammonium sulfate were all added separately to the base medium in the following amounts: 0.943 g·l−1, 3.115 g·l−1, 1.42 g·l−1, 2.0 g·l−1, and 2.0 g·l−1, respectively. The petri dishes were incubated at 25 °C for 14 d with a 5 mm diameter fungal disc. We measured the distance between colonies[20, 21].
Ethical approval
-
This study was approved by College of Education for Pure Sciences, Department of Biology, University of Basrah (No. 3066), Iraq.
-
Cladosporium endophyticum Tibpromma and KD. Hyde[12], Fig. 1.
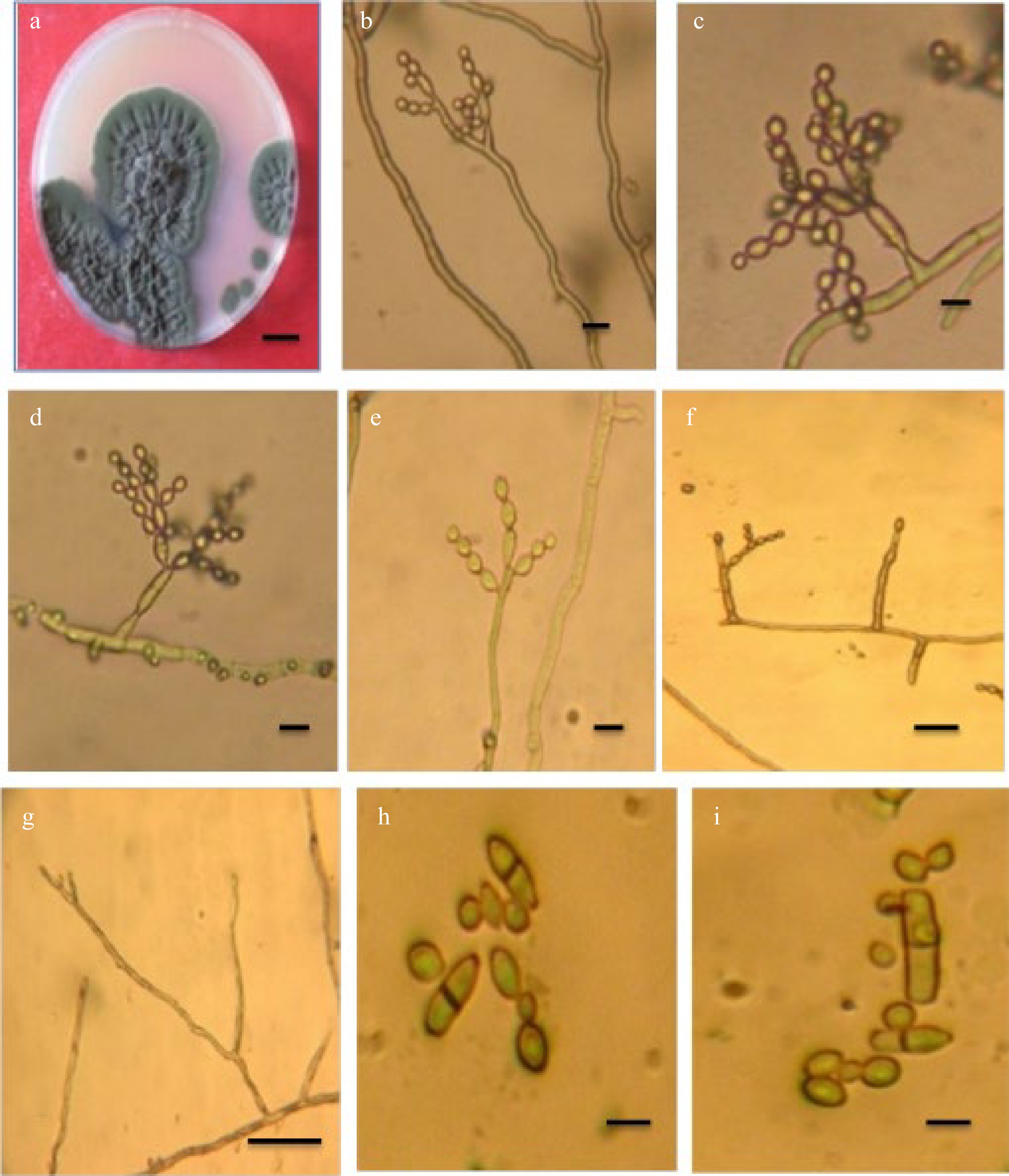
Figure 1.
Cladosporium endophyticum. (a) Colony; (b)−(e) Conidiophores with conidial chains; (f) Conidiogenous cell; (g) Conidiophore macronematocyst; (h), (i) Conidia. Scale bars (a) = 10 mm, (b)−(i) = 10 m.
Specimen examined: Iraq Basrah Province: from the air, 3 Feb 2019, coll. ZK. Abdulla
On PDA after 14 d at 25 °C, the colony diameter reached 40 mm, with an olive-green to dark olivaceous color and a wrinkled, undulating surface. The opposite was greenish-black, with a sluggish development rate and an uneven white edge.
Mycelium is superficial and immersed, septate, subhyaline, or pal olivaceous, smooth, branched, thick-walled hyphae. Conidiophores are erect, septate, usually branched, macronematous, micronematous arising single or in clusters, branched, straight, or oblique, up to 122 µm long, 2.4–3.6 µm wide. Conidiogenous cells are cylindrical, 10–25 µm long, 2.5–4 µm wide, terminal, and intercalary. Conidia hyaline to pale-olivaceous, arranged in a series, in long divider chains, smooth, little terminal conidia ovoid with rounded ends, 2.5–7.5 × 2.5–4 µm, intercalary conidia globose to ellipsoid, aseptate or one septate, 6.4–16.5 × 2.4–4.8 µm. Ramoconidia is cylindrical to ellipsoid, 0–2 septate, not constricted 10.4–27.5 × 2.5–4.5 µm.
Phylogeny
-
The alignment of ITS nucleotide sequences indicated the presence of 100% homology with no nucleic acid variations between C. endophyticum S1 in this study and C. endophyticum in GeneBank. This figure compares our sample to the NCBI database (GenBank acc. NR158360.1) and then aligns the sequences with generating these sequences.
There are 554 bp amplicon sequences among the rRNA sequences depicted in Table 1.
Table 1. Cladosporium endophyticum (NR 158360.1) has a ribosomal subunit (rsu) rRNA that was amplified using a 554 bp PCR amplicon.
Amplicon Reference locus sequences (5′ - 3′) Length rRNA TCCGTAGGTGAACCTGCGGAGGGATCATTACAAGTTGACCCCGGCCCTCGGGCCGGGATGTTCACAACCCTTTGTTGTCCGACTCTGTTGCCTCCGGGGCGACCCTGCCTCCGGGCGGGGGCCCCGGGTGGACACTTCAAACTCTTGCGTAACTTTGCAGTCTGAGTAAATTTAATTAATAAATTAAAACTTTCAACAACGGATCTCTTGGTTCTGGCATCGATGAAGAACGCAGCGAAATGCGATAAGTAATGTGAATTGCAGAATTCAGTGAATCATCGAATCTTTGAACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTTCGAGCGTCATTTCACCACTCAAGCCTCGCTTGGTATTGGGCGACGCGGTCCGCCGCGCGCCTCAAATCGACCGGCTGGGTCTTCCGTCCCCTCAGCGTTGTGGAAACTATTCGCTAAAGGGTGCCGCGGGAGGTCACGCCGCAAAACAACCCCATTTCTAAGGTTGACCTCGGATCAGGTAGGGATACCCGCTGAACTTAAGCATATCAATAAGGCGGAGGA 554 bp An rRNA subunit from Cladospoium endophyticum (NR 158360.1) was amplified using the 554 bp PCR amplicons in Fig. 2.
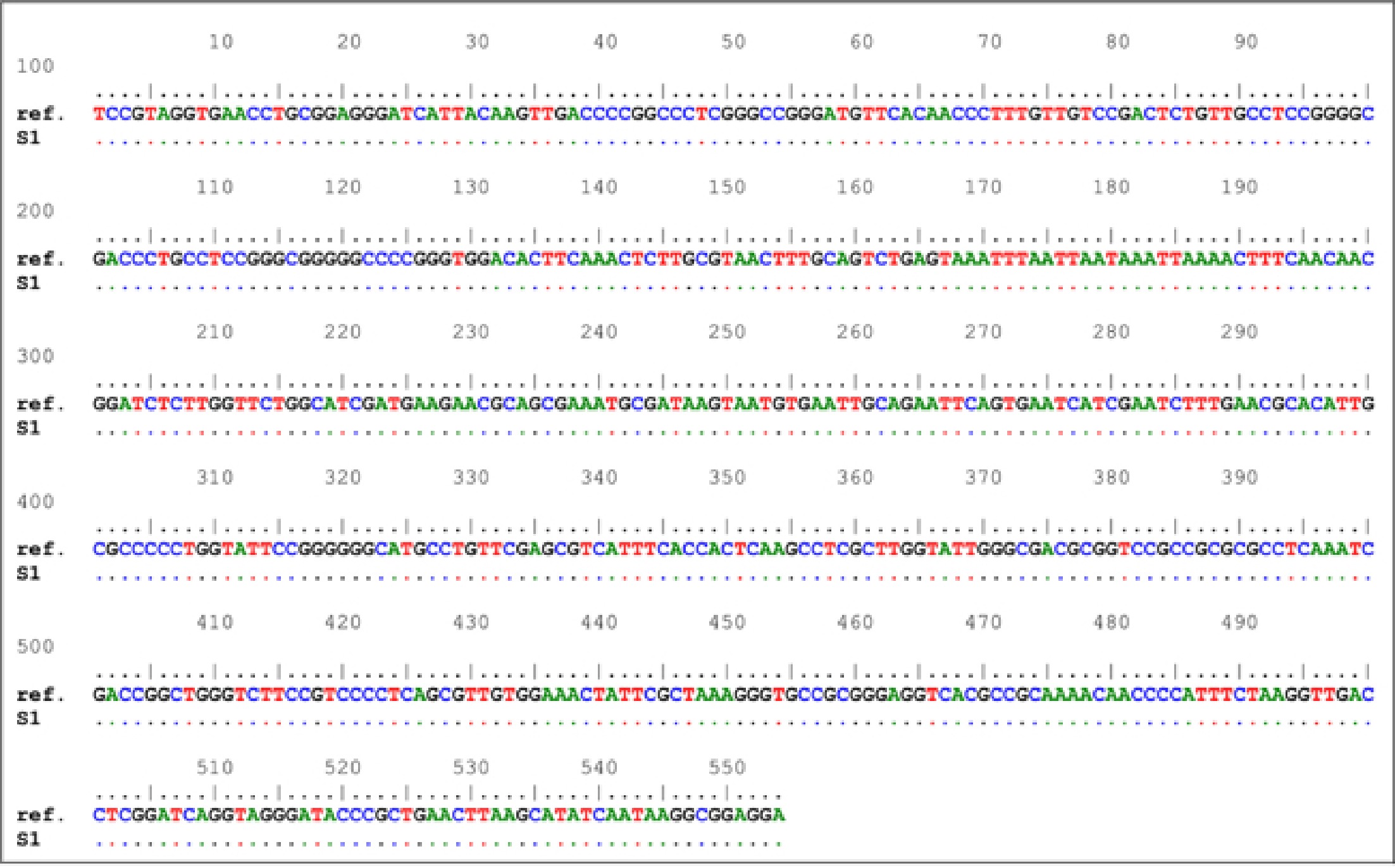
Figure 2.
Comparison of nucleic acid sequences. Cladosporium endophyticum For example, 'ref' is a reference sequence, while 'S1' is a sampling number.
Analyzed fungal samples yielded a full tree of all the fungal genomes studied. DNA sequences from other samples were also included in this phylogenetic tree, constructed using neighbor-joining techniques. Only the nucleic acid sequences from Cladosporium, a single genus, were included in the tree. It is possible to classify our three Cladosporium ribosomal sequences into three broad phylogenetic groups, which indicates that the genus' diversity in these sequences is restricted, see Fig. 3.
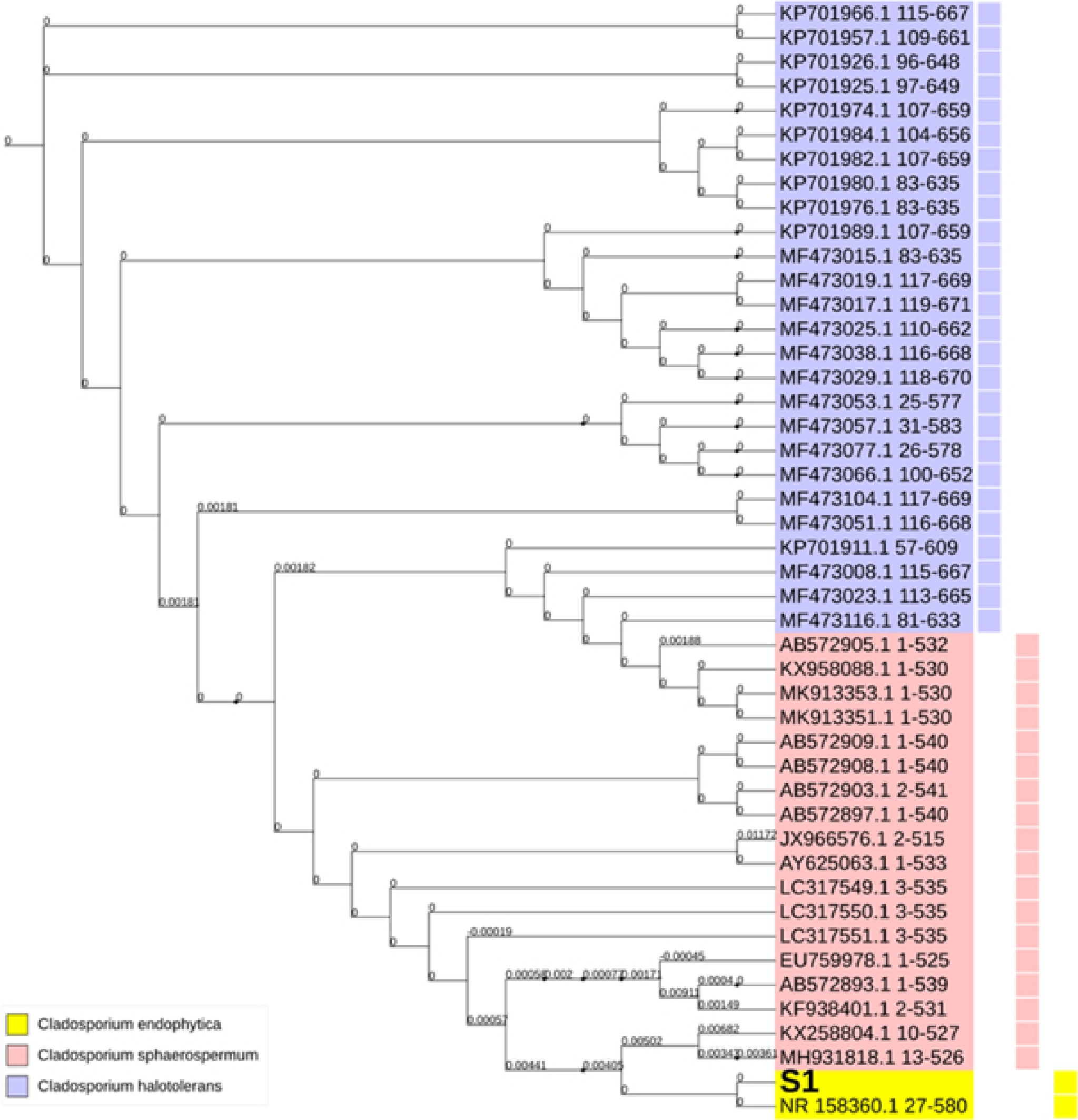
Figure 3.
Phylogenetic tree of Cladospoium endophyticum with Cladosporium sphaerospermum and Cladosporium halotolerans. Variable colors represent the variable grouping within the Genbank deposited sequences of the investigated variations. The symbol S1 denotes the code for the sample under investigation in this study.
Impact of numerous environmental factors, including temperature, pH, medium, and carbon and nitrogen supplies, on plant growth
-
For C. endophyticum, temperature had the greatest impact on radial growth (p ≤ 0.05), and RLSD was used to compare temperature and fungal growth (p ≤ 0.001). At 25 °C, fungal growth was at its best, while no growth occurred at 5 and 40 °C (Table 2).
Table 2. Impact of different temperatures on radial growth of C. endophyticum grown on PDA medium for 14 d.
Temperature Mean Std. deviation 5 0 0 10 8.66 0.57 15 17.33 1.52 20 38 1 25 46 2 30 28.66 1.15 35 6.33 1.15 40 0 0 RLSD = 1.698 (P ≤ 0.05) Table 3 showed a significant impact on various pH on radial growth of C. endophyticum, the fungus can grow at a range of pH (4−8), and pH 6 was most suitable for growth.
Table 3. Impact of different pH values on radial growth of C. endophyticum grown on PDA medium at 25°C for 14 d.
pH Mean Std. deviation 4 21.66 1.52 5 34 1 6 39.33 0.57 7 28.33 1.52 8 18.33 1.52 RLSD = 2 (P ≤ 0.05) Results showed an important impact (P ≤ 0.05) between different culture mediums and C. endophyticum, PDA, and MEA medium were more favorable, and there is no significant effect between SNA and OA media (Table 4).
Table 4. Impact of different media on radial growth of C. endophyticum incubated at 25 °C for 14 d.
Media Mean Std. deviation PDA 44.66 1.52 MEA 42 1 OA 32.33 2.08 SNA 30.66 1.52 RLSD = 2.5 (P ≤ 0.05) The difference between C. endophyticum radial growth on different carbon sources is shown in Table 5. RLSD is used to compare them. Sucrose and glucose appear significantly different from xylose, sorbitol, and manitol.
Table 5. Impact of various carbon sources on radial growth of C. endophyticum incubator at 25 °C for 14 d.
Carbon source Mean Std. deviation Sucrose 45.33 0.57 Glucose 44.00 1 Xylose 32.33 1.15 Sorbitol 27.66 0.57 Mannitol 25.00 1 RLSD = 1.39 (P ≤ 0.05) The results showed the significant difference between C. endophyticum and nitrogen sources. Ammonium nitrate was the best nitrogen source for growth, followed by ammonium phosphate, urea, and potassium nitrate, which were favorable for fungal development (Table 6).
Table 6. Impact of various nitrogen sources on radial growth of C. endophyticum at 25 °C for 14 d.
Nitrogen source Mean Std. deviation Ammonium nitrate 45.66 1.15 Ammonium phosphate 39.66 0.57 Ammonium sulphate 37.00 2 Urea 32.66 2.08 Potasium nitrate 32.33 0.57 RLSD = 2.3 (P ≤ 0.05) -
Cladosporium is a diverse genus with various genetic and phenotypic variations. C. cladosporioides, C. herbarum, and C. sphaerospermum are the three main subspecies of Cladosporium, and the majority of Cladosporium species return to the C. cladosporioides subgroup.
Morphology, genetics, and cultural characteristics have been used in most investigations to define genus and species boundaries due to Cladosporium significant morphological similarities across related species[5,6].
Cladosporium endophyticum was initially isolated from Pandanus sp. as an endophytic fungus in Thailand[12] and deposited in GenBank under the name C. endophytic, notice C. endophyticum. In this study, C. endophyticum was isolated from the air in Basrah, Iraq, and identification was based on morphological characters and genetic analysis using the ITS sequence. C. endophyticum in this study resembled that described by Tibpromma et al.[12], but there are some differences in measurements, small conidia usually ovoid with rounded end 2.5–7.5 × 2.5–4 µm, intercalary conidia globose 6.4–16.5 × 2.4–4.8 µm, ramoconidia cylindrical 10.4–27.5 × 2.5–4.5 µm. The information about this species is little because it was discovered only recently. Tibpromma et al.[12], found that C. endophyticum is sister to C. halotolerans based on phylogenetic analysis of multigene loci. However, the morphology of C. endophyticum differs from C. halotolerans by having pale-olivaceous to hyaline, globose to ovoid conidia, while C. halotolerans has subglobose to globose verrucose, brown to dark brown conidia, its ends are narrow[22]. C. endophyticum, which included the S1 sample under investigation, C. sphaerospermum, and C. halotolerans were close relatives. However, it was observed that C. sphaerospermum resided near C. endophyticum sequences. This observation entailed a tighter relationship between C. endophyticum and C. sphaerospermum sequences than with C. halotolerans. Furthermore, extremely small phylogenetic distances were observed among the incorporated organisms, which further indicates the presence of low diversity among these sequences. Within the C. endophyticum phylogenetic group, S1 resided in the immediate vicinity of the GenBank accession number NR_158360.1, which belonged to a Thai strain of C. endophyticum sequences. In contrast to both C. sphaerospermum and C. halotolerans, there is no wide spread of the C. endophyticum sequences among the deposited database in NCBI. Nevertheless, the extreme similarity between S1 and the C. endophyticum strain isolated from Thailand may indicate that our investigated sample originated from an Asian source. Accordingly, a high level of homology was derived from the currently observed fundamental similarity of the nucleic acid sequences between S1 and these Asian sequences.
The effect of environmental factors on fungal growth was studied on solid medium. The fungal growth did not depend only on physiological factors but also on the type of Cladosporium species. The temperature was the important factor that affected growth, C. endophyticum grow between 5 to 40 °C the growth decreases under 15 °C and stops above 35 °C, and 25 °C was the best temperature suited for growth. This result is in agreement with the findings of Alhussaini et al. & Gupta et al.[18, 23]. C. endophyticum can grow at a wide range of pH from 4 to 8, and the suitable pH for the growth of most fungi was 5 - 6.5, and this depended on enzymatic systems and vitamins and minerals necessary to the cell[24]. This fungus was able to grow on different carbon sources, and the highest level of mycelium growth was characterized on sucrose and glucose then xylose, sorbitol, and minnitol both came last, the simple carbon sources are consumed directly while the complex must be transformed to simple before being used by the fungus. The fungus's ability to assimilate different carbon sources gave indicators about its capacity of dominance in the ecosystem.
Nitrogen is an important factor for fungal growth. Its effect on cell function and protein production. In this study, C. endophyticum can grow on different nitrogen sources, ammonium nitrate was the best one then ammonium sulfate, and the growth was weak on urea and phosphate sulfate
-
Cladosporium is a complex genus of hyphomycetes. In this study, C. endophyticum represents a new addition for the Iraqi mycoflora, obtained from the air and identified depending on morphology and molecular characteristics and study the physiological factor effect on growth, temperature, pH, different media, carbon and nitrogen sources. The results indicated they all had significant sources of variation in mycelium radial growth.
-
The author declares that there is no conflict of interest.
- Copyright: © 2023 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Abdulla ZK. 2023. Taxonomy and biology of Cladosporium endophyticum as the first record in Iraq. Studies in Fungi 8:5 doi: 10.48130/SIF-2023-0005
Taxonomy and biology of Cladosporium endophyticum as the first record in Iraq
- Received: 05 December 2022
- Accepted: 04 January 2023
- Published online: 02 February 2023
Abstract: Cladosporium species are cosmopolitan organisms. Conidiogenous loci distinguish this genus. Their spores can be isolated from different sources. The morphological analysis and ITS sequence were used to identify Cladosporium endophyticum isolated from air. The findings demonstrate that C. endophyticum S1 and C. endophyticum Thailand strain were almost identical in this investigation. The C. endophyticum strain is isolated from Thailand, and it has a distinctive phylogenetic clade. This clade resided in the vicinity of Cladosporium sphaerospermum and Cladosporium halotolerans, respectively. In order to study the physiological factor effect on C. endophyticum on solid media, different temperatures were used (5, 10, 15, 20, 25, 30, 35, 40), pH (4, 5, 6, 7), media malt extract agar (MEA), potato dextrose agar (PDA), oatmeal agar (OA), synthetic nutrient-deficient agar (SNA), carbon (sucrose, glucose, sorbitol, xylose, manitol), nitrogen (ammonium nitrate, ammonium sulphat, urea, ammonium phosphate, potassium nitrate). The results showed all factors' highly significant (p ≤ 0.05) effect. The faster growth was found at 25 °C, pH suitable for growth 5 and 6. The maximum radial growth was found on PDA and MEA. The fungus C. endophyticum can utilize all carbon sources, and faster growth was recorded on sucrose, then glucose, xylose, sorbitol, and manitol. The fungus grows on all nitrogen sources, and mycelium growth was good on ammonium nitrate, ammonium phosphate, ammonium sulfate, urea, and potassium nitrate. Cladosporium endophyticum is newly recorded from Basrah, Iraq, and it has been deposited in GenBank with accession numbers for nucleotide sequence (MW298524).
-
Key words:
- Cladosporium /
- Morphology /
- ITS sequence /
- Taxonomy.












