-
As a perennial warm-season grass, Zoysia (Zoysia spp.) is recognized for its low maintenance requirements as well as relatively high tolerance to drought, disease, and traffic, and is widely cultivated for athletic fields, home lawns, and other recreational sites, particularly in east Asia[1]. With the rapid development of the turf industry, it is extensively recognized that germplasm resources are hugely important. Therefore, a large number of researches are focussed on the collection[2−5], evaluation[6−8], and breeding[9−11] of Zoysia spp.
Zoysia species exhibit a high rate of outcrossing and are prone to interspecific hybridization, resulting in a wide range of genetic variation among Zoysia plants[12]. Kimball et al.[13] identified interspecific hybridization and hypothesized that these hybrids were the result of introgression between species through common breeding methods of Zoysia, including directed hybridization of selected parents and cross-pollination in open cross areas. Genetic background analysis of abundant germplasm is an important prerequisite for identifying parents and breeding Zoysia varieties[14]. Traditionally morphological identification has several drawbacks due to the susceptibility to environmental factors and plant growth period as well as the limited morphological indexes. Furthermore, some germplasm are difficult to differentiate based on phenotype alone[15]. Molecular markers are an effective means to grasp the genetic information of germplasm. Among several common molecular markers, simple sequence repeats (SSRs) are widely used molecular markers in plant genetics and breeding, due to their multiallelic, codominant inheritance and extensive genome coverage[16]. Sequence-related amplified polymorphism (SRAP) was developed by Li & Quiros[17] in Brassica. This method is advantageous due to its simplicity, reasonable throughput rate, disclosure of numerous codominant markers, and targeting of open reading frames (ORFs)[18]. Both marker systems have been applied to a range of fields, including the analysis of genetic diversity of germplasm[19−22], the identification of cultivar and marker-trait association[23]. SSR and SRAP have been reported to be effectively utilized to examine the genetic diversity[4], analyze genetic similarity among cultivars[24], and identify molecular markers linked with quantitative trait loci for biotic and abiotic stress tolerance in Zoysia[3, 25]. These markers also serve as a potent tool for constructing DNA fingerprints[7, 26, 27].
The evaluation and improvement of germplasm are of great significance for the effective utilization of these resources. Previous researchers investigated the genetic variation of some Zoysia germplasm. Anderson[28] measured inflorescence traits, morphological characteristics, and restricted fragment length polymorphisms (RFLPs) to evaluate the genetic and morphological variations in 11 species of Zoysia. Li et al.[29] reported a linkage map of Zoysia matrella based on SSR markers, combined the previously reported SSR linkage maps of different Zoysia species (Z. japonica and Z. matrella) and different mapping populations (F1, S2, and F2)[30] to construct a complete SSR genetic linkage map. Xie et al.[31] conducted a study to analyze the genetic diversity and relationships of 84 Zoysia germplasms using inter-simple sequence repeat (ISSR) markers. Moore et al.[32] analyzed changes in levels of allelic diversity at the gene and population levels in 40 Zoysia cultivars released between 1910 and 2016 using SSR markers.
In this study, 45 Chinese Zoysia germplasm collections were analyzed by SSR and SRAP markers. The results of these analyses were used to clarify the genetic relationship, analyze the genetic diversity of 45 Chinese Zoysia germplasm collections, and construct the fingerprint to distinguish breeding lines from the two commercially available cultivars at the molecular level. The aim of this study was to provide a theoretical basis for the development, evaluation, and breeding of Zoysia germplasm.
-
Forty-five Chinese Zoysia germplasm collections were used for the study, among which 38 were collected from the provinces of Liaoning, Anhui, Zhejiang, and Hainan in China, five were breeding lines from radiation mutagenesis and two commercial Z. japonica cultivars, 'Lanyin No. 3' and 'Qingdao' (Table 1). Forty-five Zoysia germplasm collections were grown in the resource nursery of Turf Experiment Station of Nanjing Agricultural University, located at 119°14′38″ east longitude and 31°49′46″ north latitude, with a subtropical monsoon climate and an average annual temperature of 15.2 °C. A randomized block design was utilized with each germplasm planted in an area of 3 m × 3 m with three replicates and plots spaced 2 m apart in August, 2020. About 100 stolons were planted evenly in each plot with 2−3 stem nodes for each stolon. Plants were watered twice weekly and fertilized once a month. Plants were not trimmed in 2021 in order to measure morphological indexes, and trimmed once a week in 2022 to evaluate turf density.
Table 1. List of Chinese Zoysia germplasm collections used in this study.
No. Germplasm ID Type Species Source 1 ZG003 Breeding material Z. sp. Liaoning 2 ZG004 Breeding material Z. sp. Anhui 3 ZG007 Breeding material Z. sinica Anhui 4 ZG008 Breeding material Z. sinica Radiation mutagenesis
(parent from Anhui)5 ZG009 Breeding material Z. matrella Jiangsu 6 ZG011 Breeding material Z. pacifica Zhejiang 7 ZG013 Breeding material Z. sp. Anhui 8 ZG015 Breeding material Z. sinica Anhui 9 ZG017 Breeding material Z. sp. Anhui 10 ZG018 Breeding material Z. sp. Anhui 11 ZG021 Breeding material Z. sp. Anhui 12 ZG022 Breeding material Z. sp. Anhui 13 ZG023 Breeding material Z. matrella Anhui 14 ZG025 Breeding material Z. sp. Anhui 15 ZG026 Breeding material Z. matrella Anhui 16 ZG028 Breeding material Z. sp. Anhui 17 ZG029 Breeding material Z. sp. Zhejiang 18 ZG030 Breeding material Z. matrella Zhejiang 19 ZG032 Breeding material Z. sp. Zhejiang 20 ZG035 Breeding material Z. sp. Zhejiang 21 ZG037 Breeding material Z. sp. Zhejiang 22 ZG038 Breeding material Z. sp. Zhejiang 23 ZG040 Breeding material Z. sp. Zhejiang 24 ZG041 Breeding material Z. sp. Anhui 25 ZG043 Breeding material Z. sp. Anhui 26 ZG044 Breeding material Z. sp. Anhui 27 ZG046 Breeding material Z. sinica Hainan 28 ZG047 Breeding material Z. sp. Hainan 29 ZG048 Breeding material Z. pacifica Hainan 30 ZG049 Breeding material Z. sp. Hainan 31 ZG050 Breeding material Z. matrella Hainan 32 ZG053 Breeding material Z. pacifica Hainan 33 ZG056 Breeding material Z. pacifica Anhui 34 ZG057 Breeding material Z. sp. Anhui 35 ZG058 Breeding material Z. matrella Anhui 36 ZG059 Breeding material Z. sinica Radiation mutagenesis
(parent from Anhui)37 ZG060 Breeding material Z. sp. Anhui 38 ZG061 Breeding material Z. sp. Anhui 39 ZG062 Breeding material Z. sp. Anhui 40 ZG063 Breeding material Z. sp. Anhui 41 ZG081 Breeding material Z. japonica Radiation mutagenesis
(parent from Anhui)42 ZG082 Breeding material Z. japonica Radiation mutagenesis
(parent from Anhui)43 ZG083 Breeding material Z. japonica Radiation mutagenesis
(parent from Anhui)44 Lanyin
No. 3Cultivar Z. japonica Gansu 45 Qingdao Cultivar Z. japonica Shandong Morphological characteristic data collection
-
Morphological data were collected on the 15th in June and September 2021. Six morphological characteristics of 45 Chinese Zoysia germplasm collections were determined as follows: leaf length and leaf width were obtained by randomly measuring the length and middle width of the third fully expanded leaf from the top, respectively. Measurements were repeated 10 times and the average was calculated as the final value for each replicate. Ten healthy stolons were randomly selected and the length and diameter of the fourth section were measured as internode length and stem diameter, respectively. The turf height, which is the natural height of plant growth, was measured using the five-point method[33]. Turf density was determined by counting the number of tillers in a 5 cm × 5 cm wire frame and each plot was counted three times.
Genomic DNA extraction
-
Genomic DNA was extracted from young Zoysia leaves (0.1 g) using the modified CTAB method[34]. The quality of DNA was verified by 0.8% agarose gel electrophoresis, and the DNA concentration was determined by NanoReady. The DNA of 45 samples was diluted to 50 ng·μL−1 using purified water and stored in the refrigerator at 4 °C or −20 °C for later use.
SSR and SRAP analysis
-
Forty SSR primer pairs from Röder et al.[35] and Tsuruta et al.[36] were used in this study. SSR-PCR and SRAP-PCR was performed in a total volume of 20 μL containing 1 μL genomic DNA, 10 μL of 2 × Mix (Yeasen Biotechnology Co., Ltd., Shanghai, China), 1 μL of 10 μmol·L−1 each PCR primer and 7 μL purified water. SSR-PCR reactions were performed in a Bio-Rad thermal cycler (Bio-Rad Inc., Hercules, CA, USA), DNA amplifications were performed with an initial step at 94 °C for 3 min, followed by 35 cycles of 50 s at 94 °C, 30 s at 55 °C, 1 min at 72 °C, and a 10 min final extension step at 72 °C.
According to the SRAP primer design method published by Li & Quiros[17], 10 forward primers and five reverse primers were randomly selected. SRAP-PCR reactions were performed with an initial step at 94 °C for 4 min and five cycles of 60 s at 37 °C and 60 min at 72 °C. Then 35 cycles of denaturation at 94 °C for 1 min, annealing at 50 °C for 1 min, and extension at 72 °C for 10 s; and then a final extension at 72 °C for 10 min.
The amplifications were performed in Applied Biosystems Veriti® thermal cycler. Amplification products were stored at 4 °C before being electrophoresed through 10% non-denatured polyacrylamide gels in 1 × TBE (pH 8.0) buffer running at 120 V constant voltage for 1.5 h and then the gels were stained with fast silver stain[37, 38]. According to the silver staining results, 10 SSR primer pairs and nine SRAP primer pairs were initially screened against 45 Chinese Zoysia germplasm collections (Tables 2 & 3).
Table 2. Simple sequence repeats (SSR) primer sequences used for studying genetic diversity of 45 Chinese Zoysia germplasm collections.
Primer name Forward primer (5'-3') Reverse primer (5'-3') M3A10 CGAACGCGACATGACAATC TCATGATGTTGGCAACCAC Xgwm37-7D ACTTCATTGTTGATCTTGCATG CGACGAATTCCCAGCTAAAC Xgwm102-2D TCTCCCATCCAACGCCTC TGTTGGTGGCTTGACTATTG Xgwm111-7D TCTGTAGGCTCTCTCCGACTG ACCTGATCAGATCCCACTCG Xgwm120-2B GATCCACCTTCCTCTCTCTC GATTATACTGGTGCCGAAAC Xgwm169-6A ACCACTGCAGAGAACACATACG GTGCTCTGCTCTAAGTGTGGG Xgwm445-2D GTTGAGCTTTTCAGTTCGGC ACGGAGAGCAACCTGCC Xgwm46-7B GCACGTGAATGGATTGGAC TGACCCAATAGTGGTGGTCA Xgwm52-3D CTATGAGGCGGAGGTTGAAG TGCGGTGCTCTTCCATTT Xgwm234-5B GAGTCCTGATGTGAAGCTGTTG CTCATTGGGGTGTGTACGTG Table 3. Sequence-related amplified polymorphism (SRAP) primer sequences used for studying genetic diversity of 45 Chinese Zoysia germplasm collections.
Forward primer (5'-3') Me1 TGAGTCCAAACCGGATA Me2 TGAGTCCAAACCGGAGC Me3 TGAGTCCAAACCGGACC Me4 TGAGTCCAAACCGGACA Me5 TGAGTCCAAACCGGTGC Me6 TGAGTCCAAACCGGAGA Me7 TGAGTCCAAACCGGACG Me8 TGAGTCCAAACCGGAAA Me9 TGAGTCCAAACCGGAAC Me10 TGAGTCCAAACCGGAAT Reverse primer (5'-3') Em1 GACTGCGTACGAATTCAA Em2 GACTGCGTACGAATTCTG Em3 GACTGCGTACGAATTGAC Em4 GACTGCGTACGAATTTGA Em5 GACTGCGTACGAATTAAC Data analysis
-
Gel images from all accessions were visually evaluated and coded as either a '1' for the presence or '0' for the absence of a band for each marker. Based on this method, matrices of '0' and '1' were obtained for 45 germplasm collections based on the amplification of each pair of primers. Popgene 3.2 software was used to calculate the number of alleles (Na), effective number of alleles (Ne), Shannon information index (I) and Nei's gene diversity (H). Genetic distances were calculated for the 45 Chinese Zoysia germplasm collections according to Dice[39] using NTSYS-pc. Genetic similarity coefficient (GS) was calculated, and dendrograms was constructed using unweighted pair-group method with arithmetic averages (UPGMA) by the NTSYS-pc computer program package. The confidence probability of the fingerprint was calculated based on the probability formula P = 1/2n, where n is the number of alleles, i.e., the number of polymorphic bands for each primer pair.
-
A total of 395 bands were amplified with 19 pairs of primers, among which 380 were polymorphic bands, with a polymorphism ratio of 96.20% (Table 4). Ten SSR primer combinations amplified a total of 231 polymorphic bands with a polymorphism ratio of 97.18%. The number of bands scored per SSR primer combination ranged from 16 to 36, with a mean of 23.70. A total of 149 polymorphic bands were amplified by nine pairs of SRAP primers, and the polymorphism ratio was 93.43%. The number of bands scored per SRAP primer combination ranged from 13 to 30, with a mean of 17.56. Based on the amplification of SSR and SRAP primers, the effective alleles ranged from 1.192 to 1.556 with an average of 1.332, Shannon information index ranged from 0.236 to 0.510 with an average of 0.350, and Nei's gene diversity index ranged from 0.146 to 0.338, and the mean value is 0.219. The results showed that there was a high level of genetic diversity among the tested germplasm collections, and the SSR and SRAP primers screened were suitable for amplification detection of Zoysia.
Table 4. Polymorphism results from amplification by simple sequence repeats (SSR) and sequence-related amplified polymorphism (SRAP) primers in 45 Chinese Zoysia germplasm collections.
Primer Total number of
amplified bandsNumber of
polymorphic bandsPercentage of
polymorphic bands (%)Effective number
of allelesShannon
information indexNei's diversity
indexM3C06 27 27 100.00 1.443 0.426 0.272 Xgwm37-7D 17 16 94.12 1.201 0.259 0.146 Xgwm102-2D 24 22 91.67 1.350 0.352 0.224 Xgwm111-7D 28 28 100.00 1.254 0.295 0.172 Xgwm120-2B 36 35 97.22 1.556 0.510 0.338 Xgwm169-6A 16 16 100.00 1.276 0.333 0.197 Xgwm445-2D 32 32 100.00 1.396 0.398 0.250 Xgwm46-7B 16 15 93.75 1.394 0.399 0.255 Xgwm52-3D 20 19 95.00 1.409 0.413 0.262 Xgwm234-5B 21 21 100.00 1.249 0.305 0.179 SSR marker average 23.70 23.10 97.18 1.353 0.369 0.229 Me3-Em1 30 30 100.00 1.275 0.337 0.199 Me2-Em2 17 16 94.12 1.314 0.333 0.204 Me3-Em2 13 12 92.31 1.448 0.389 0.258 Me4-Em4 20 20 100.00 1.422 0.417 0.264 Me6-Em4 14 13 92.86 1.192 0.236 0.185 Me5-Em2 13 11 84.62 1.252 0.292 0.174 Me3-Em3 19 17 89.47 1.410 0.388 0.251 Me6-Em2 17 16 94.12 1.217 0.286 0.163 Me7-Em1 15 14 93.33 1.254 0.284 0.171 SRAP marker average 17.56 116.56 93.43 1.309 0.329 0.208 Average of all markers 20.79 20 96.20 1.332 0.350 0.219 Total 395 380 — — — — Genetic similarities
-
NTSYS-pc software was used to calculate the value of GS between any two materials according to Dice's coefficient, which was based on the combined amplification results of SSR and SRAP (Table 5). The GS of the 45 germplasm collections ranged from 0.623 to 0.856, with an average value of 0.727 and a variation of 0.233. ZG048 was most genetically similar to ZG053 (GS = 0.856), and ZG025 was most genetically dissimilar to ZG032 (GS = 0.632). Furthermore, two commercial cultivars, Lanyin No. 3 and Qingdao, were found to be highly genetically similar (GS = 0.815). The least genetically similar to Lanyin No. 3 was ZG040 (GS = 0.651), while the least genetically similar to Qingdao was ZG008 (GS = 0.661).
Table 5. Genetic similarity coefficients (GS) among the 45 Chinese Zoysia germplasm collections.
Germplasm ID Highest similarity
(germplasm ID
compared to)Lowest similarity
(germplasm ID
compared to)Average ZG003 0.8532 (ZG004) 0.6228 (ZG032) 0.738 ZG004 0.8532 (ZG003) 0.6582 (ZG032) 0.756 ZG007 0.8127 (ZG009) 0.6532 (ZG029) 0.733 ZG008 0.7772 (ZG015) 0.6278 (ZG081) 0.703 ZG009 0.8127 (ZG007) 0.6633 (ZG081) 0.738 ZG011 0.7873 (ZG021) 0.6456 (ZG081) 0.716 ZG013 0.8152 (ZG021) 0.6684 (ZG081) 0.742 ZG015 0.7772 (ZG008) 0.6608 (ZG060) 0.719 ZG017 0.7722 (ZG025) 0.6582 (ZG032) 0.715 ZG018 0.8101 (ZG021) 0.6557 (ZG032) 0.733 ZG021 0.8304 (ZG022) 0.6658 (ZG046) 0.748 ZG022 0.8304 (ZG021) 0.6759 (ZG032) 0.753 ZG023 0.7848 (ZG026) 0.6532 (ZG081) 0.719 ZG025 0.7722 (ZG017) 0.6228 (ZG032) 0.698 ZG026 0.7848 (ZG023) 0.6430 (ZG032) 0.714 ZG028 0.7772 (ZG021) 0.6684 (ZG060) 0.723 ZG029 0.7646 (ZG035) 0.6506 (ZG081) 0.708 ZG030 0.7823 (ZG043) 0.6582 (ZG081) 0.720 ZG032 0.7418 (ZG037) 0.6228 (ZG025) 0.682 ZG035 0.8329 (ZG037) 0.6684 (ZG081) 0.751 ZG037 0.8329 (ZG035) 0.6532 (ZG081) 0.743 ZG038 0.8304 (ZG037) 0.6658 (ZG081) 0.748 ZG040 0.7316 (ZG047) 0.6456 (ZG003) 0.689 ZG041 0.7873 (ZG043) 0.6633 (ZG082) 0.725 ZG043 0.7873 (ZG041) 0.6658 (ZG032) 0.727 ZG044 0.7722 (ZG035) 0.6481 (ZG082) 0.710 ZG046 0.7519 (ZG047) 0.6456 (ZG081) 0.699 ZG047 0.7899 (ZG048) 0.6759 (ZG081) 0.733 ZG048 0.8557 (ZG053) 0.6633 (ZG081) 0.760 ZG049 0.7873 (ZG047) 0.6810 (ZG026) 0.734 ZG050 0.7595 (ZG058) 0.6709 (ZG004) 0.715 ZG053 0.8557 (ZG048) 0.6684 (ZG032) 0.762 ZG056 0.7873 (ZG057) 0.6684 (ZG103) 0.728 ZG057 0.8051 (ZG059) 0.6633 (ZG082) 0.734 ZG058 0.7873 (ZG013) 0.6532 (ZG082) 0.720 ZG059 0.8051 (ZG057) 0.6785 (ZG032) 0.742 ZG060 0.7646 (ZG049) 0.6354 (ZG032) 0.700 ZG061 0.8152 (ZG062) 0.6506 (ZG025) 0.733 ZG062 0.8253 (ZG063) 0.6557 (ZG032) 0.741 ZG063 0.8253 (ZG062) 0.6608 (ZG046) 0.743 ZG081 0.7949 (ZG083) 0.6278 (ZG032) 0.711 ZG082 0.7899 (ZG083) 0.6354 (ZG025) 0.713 ZG083 0.7949 (ZG081) 0.6481 (ZG040) 0.722 Lanyin No. 3 0.8152 (ZG105) 0.6506 (ZG040) 0.733 Qingdao 0.8152 (ZG103) 0.6608 (ZG008) 0.738 Cluster analysis
-
Based on the results of SSR and SRAP molecular markers, the Dice genetic similarity coefficients were used to cluster 45 Chinese Zoysia germplasm collections. The cophenetic correlation for the UPGMA clustering was high (r = 0.75), suggesting that the cluster analysis strongly represented the similarity matrix. A UPGMA cluster was constructed based on combining data from both markers, which divided the 45 accessions into six major clusters at a similarity index value of 0.71 (Fig. 1). Cluster I contained 32 germplasm collections, comprised of 17 germplasm collections from Anhui, six germplasm collections from Zhejiang, five germplasm collections from Hainan, one material from Liaoning, one radiation mutagenic material and two commercial cultivars. Cluster II comprised of three germplasm collections from Anhui. Cluster III consisted of four accessions, comprising of one from Jiangsu, two from Anhui and one breeding germplasm collections. Cluster IV included two germplasm collections from Zhejiang. Cluster V included only one germplasm from Hainan, ZG046, indicating that ZG046 had some genetic differences from other germplasm collections from the same region. Cluster VI contained three radiation mutagenic materials. Using a genetic similarity coefficient of 0.73, the samples of Cluster I were differentiated into three subgroups: A, B, and C. Subgroup A consisted of Lanyin No. 3, Qingdao, and five germplasm collections from Anhui and Liaoning. Subgroup B contained 10 from Zhejiang and Anhui and one from Hainan (ZG050). Subgroup C included one radiation mutagenic material, nine germplasm collections from Zhejiang and Anhui, and four germplasm collections from Hainan.
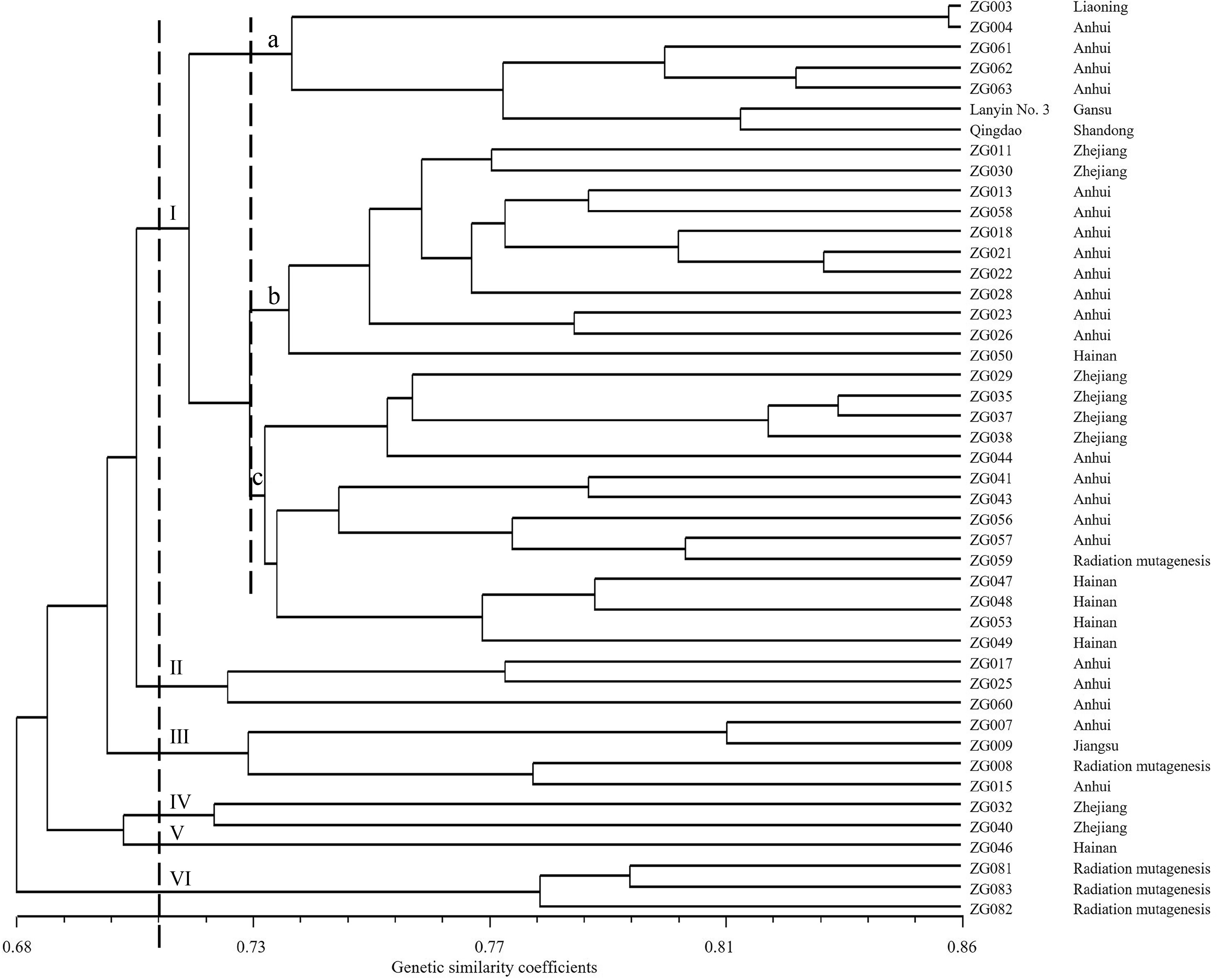
Figure 1.
Unweighted pair-group method with arithmetic averages dendrogram of the 45 Chinese Zoysia germplasm collections constructed using simple sequence repeats (SSR) and sequence-related amplified polymorphism (SRAP) markers.
Cluster I contained most of the germplasm collections and had the most diverse source locations, including four sources, indicating that there was genetic similarity among the germplasm from different sources. For example, ZG003 from Liaoning and ZG004 from Anhui preferentially clustered together and then aggregated with the rest of the germplasm collections in Cluster I. The genetic similarity coefficient of the two germplasm collections was 0.853, which was higher than the average. Aside from Cluster I, the other accessions preferentially clustered together according to their source location, such as Clusters II, IV, and VI.
In terms of morphological characteristics of the major clusters (Table 6), germplasm collections in subgroup A of Cluster Ⅰ were mainly presented with long and wide leaves, long internodes, large diameter of above ground stems and low density. Germplasm collections of subgroup B was tall and dense, making them suitable for vegetative propagation in ecological restoration or green space construction in parks. Germplasm collections in Subgroup C and Clusters II and III exhibited intermediate morphological characteristics, belonging to the intermediate type germplasm collections. Germplasm collections from Cluster IV (ZG032 and ZG040) were tall with low density which may not be suitable for utilizing as turf. ZG046 was the sole material in Cluster V with short leaves and short internodes, superior traits for sports field turf. Germplasm collections in Cluster VI were mainly characterized by narrow leaves and fine texture, suggesting potential to be used for ornamental lawn in urban green spaces.
Table 6. Mean value of main morphological characteristics of the six clusters of 45 Chinese Zoysia germplasm collections.
Cluster/subgroup Leaf width
(mm)Leaf length
(cm)Internode length
(cm)Stem diameter
(mm)Turf height
(cm)Turf density
(tiller number cm−2)I A 4.66 10.24 5.13 1.58 11.44 4.66 B 3.42 7.83 3.59 1.35 10.55 3.42 C 3.62 6.75 4.62 1.34 14.00 3.62 II − 3.70 7.29 3.36 1.23 10.08 2.81 III − 3.74 6.80 3.82 1.37 11.03 3.03 IV − 3.66 6.66 3.72 1.41 22.96 2.17 V − 3.47 4.15 3.11 1.38 9.07 2.81 VI − 3.27 6.33 4.63 1.37 8.14 2.80 Mean 3.72 7.51 4.22 1.38 10.83 3.19 Fingerprints of 45 Zoysia germplasms
-
The selected SSR and SRAP primers were used to amplify the DNA from 45 Chinese Zoysia germplasm collections, and the results of amplified bands were stable and reproducible. Fewer primers are preferred if they are able to distinguish among varieties. In this study, one SSR primer set (Xgwm234-5B) and two SRAP primer sets (Me3-Em1 and Me3-Em2) were selected to construct SSR and SRAP fingerprints of 45 germplasm collections, respectively, by considering the clarity and percentage of polymorphic of amplified bands for each primer pair (Table 7). The primer pairs Xgwm234-5B, Me3-Em1, and Me3-Em2 amplified 21, 30, and 12 polymorphic bands, respectively. The fingerprint detection probability formula P = 1/2n was used of which n was 21 and 42 for SSR and SRAP fingerprints, respectively. The confidence probability of both SSR and SRAP fingerprints were more than 99.999%. These results indicated that the fingerprint obtained in this study could be used to distinguish among the 45 Chinese Zoysia germplasm collections.
Table 7. Fingerprints of 45 Chinese Zoysia germplasm collections generated by simple sequence repeats (SSR) and sequence-related amplified polymorphism (SRAP) markers.
No. Germplasm ID Digital DNA fingerprint SSR (Xgwm234-5B) SRAP (Me3-Em1+Me3-Em2) 1 ZG003 111000100010100110000 000001000000000001000000011011-0001101010010 2 ZG004 000001000010000100000 000000100001000001000001010010-0001101010010 3 ZG007 100000100010000100100 000000100010001000010001001110-0000001100001 4 ZG008 001000000010000100111 100000000010000000001000001100-0000001110011 5 ZG009 101010000010100101100 000001000000010100010001001110-0000001110001 6 ZG011 100000000010000100010 000000000000000001000000000000-1001101010010 7 ZG013 000000100010000100000 010001000000010001010010010000-0001101010010 8 ZG015 001000000010000100001 100010001100000000001000000100-0000001000011 9 ZG017 000100100010000100101 000010001000000000010010001100-0000101100010 10 ZG018 000000000000000100001 000001000010000000110010110010-1001101010010 11 ZG021 101000100010000100000 010001000000010001000010110001-0001101010010 12 ZG022 100000110010001100000 000001000000000000000010100000-0001101010010 13 ZG023 000000000010000000000 000010000100010001010010100100-1001111010010 14 ZG025 000000000010000101100 010010001000010000000010001110-0000101100010 15 ZG026 001001100011100100110 010001101100000001010000100100-1001111010010 16 ZG028 000001100010000110000 010001000000000010000010101010-0001101010010 17 ZG029 001000000010001100100 000010000000010000000000100100-1001101010010 18 ZG030 001001000111000100011 100010100100000001010010100000-1000111010011 19 ZG032 000001000010001110000 100010000010010001010110100001-0000001010001 20 ZG035 001010000010000100010 000010001010010000000000100010-1001101010011 21 ZG037 000001100010001100000 000010001000010000000010100000-1001101010011 22 ZG038 000000000010000100000 010010000100010000000000010000-1001101010011 23 ZG040 000000000010000100100 000010010100000000001010010000-0000101010011 24 ZG041 010010100010000100000 010010000100000001000100100000-0001101010011 25 ZG043 000000100011000100000 000000000000000000101010100011-0000101010011 26 ZG044 011100010010000100000 010001001010001000000010100000-1001101011011 27 ZG046 000000001000000110010 010000100100010000110010001000-0000111110011 28 ZG047 000000000000000100010 000000000001010000000010101000-0000101110011 29 ZG048 001001110010001101010 010010000001010001001000001000-0001111110011 30 ZG049 100000000010000111110 100010000100010100001010001000-0000101110011 31 ZG050 000001000010000100011 010010000000110001010000000000-1001111010011 32 ZG053 001000000011000101010 010010000001010001001000001000-0001111110010 33 ZG056 000001000010010100010 010000001100000000010010001010-0000111110011 34 ZG057 001000100010101100010 010001000000010010010010001010-0000001110011 35 ZG058 001001100010000100110 010001000010010000010010001000-1000111010011 36 ZG059 010000000010000100010 010001000000010000000001001010-0000101110001 37 ZG060 000000000010000100110 001001000100110000010000001000-0000101110001 38 ZG061 000001100010000100000 000000100001000000100000000001-1001101010001 39 ZG062 100001100010000100101 000100100001001000100001010000-0001101010001 40 ZG063 000000000011000100100 000000100001010000100001010110-0001101010101 41 ZG081 010001000010010100110 000100000000100100010010101100-0011101010101 42 ZG082 001000010010000000110 000100010000100100010001100101-0011101010100 43 ZG083 000001000010100100110 000100000000100100010000101100-0011101010101 44 Lanyin No. 3 000000000000100000000 000000000000100100000001100001-0001011001001 45 Qingdao 110001000010000101000 000000000000000100010000101100-1001001010001 DNA from 45 Chinese Zoysia germplasm collections were amplified with primers Xgwm234-5B, Me3-Em1, and Me3-Em2. The presence or absence of bands in the same location was transformed into the corresponding digital information of 1 or 0, respectively, to form the digital fingerprints. -
Many Zoysia species, such as Z. sinica, Z. japonica, and Z. matrella, were utilized as turfgrass with relatively substantial genetic diversity. Weng et al.[40] analyzed 131 Zoysia plants collected from Taiwan with random amplified polymorphic DNA (RAPD) and isoenzymes and determined that these germplasms exhibit high genetic variation at the DNA level. Kimball et al.[41] selected 50 pairs of SSR primers to amplify 62 DNA samples from Zoysia cultivars and accessions, and the genetic similarity obtained from their analysis ranged from 0.29 to 0.51. In our study, the genetic similarity coefficient of 45 germplasm collections ranged from 0.623 to 0.856 indicating that the diversity of 45 germplasm collections was lower. The level of genetic diversity and similarity within and between natural populations is determined by the interaction of climate differences and gene flow, with climate being a major driving force for organisms to adapt to the environment and generate heritable mutations[42]. Zoysia has gained popularity and been widely utilized around the world due to its remarkable heat tolerance, cold tolerance, and saline-alkali tolerance, which results in the complex genetic variation of Zoysia plants.
In this study, the 45 germplasm collections were divided into six clusters by UPGMA. The geographic origin of Cluster I was relatively complex, comprising of germplasm collections from Anhui (17 samples), Zhejiang (six samples), Hainan (five samples), Liaoning (one sample), one radiation mutagenic material, and two commercial cultivars. Notably, the two control cultivars of Lanyin No. 3 and Qingdao are both in subgroup A of cluster I, yet their origins are quite distinct. Lanyin No. 3 was introduced from the United States by Gansu Ecology Institute in 1988, while Qingdao was collected in Jiaozhou Bay, Shandong Province in 1990, and subsequently cultivated and domesticated. Similarly, it has been reported that the Zoysia cultivars Empire, JaMur, and Atlantic were difficult to be differentiated by 40 SSR markers[24]. Numerous warm-season turfgrass cultivars are highly genetically similar despite reported pedigrees or place of collection[43, 44]. Utilizing other types of markers such as single nucleotide polymorphisms (SNPs) or whole genome sequencing of these cultivars might be able to provide additional information to uncover genotypic diversity. Cluster V was comprised of a single ZG046 from Hainan, while the other five (ZG047, ZG048, ZG049, ZG050, and ZG053) from Hainan were placed in Cluster I. Additionally, these five germplasm collections exhibited significant differences from ZG046 in leaf length and leaf width. Results showed that the genetic distance between ZG046 and the other five germplasm collections was significant. Therefore, ZG046 can be used as a parent material to expand the genetic basis of the breeding population and increase the genetic diversity of breeding material, which would be beneficial for further Zoysia breeding improvement.
In this study, 45 Chinese Zoysia germplasm collections were characterized and analyzed based on their morphological traits and molecular clustering results. The morphological traits of Zoysia were found to be influenced by various factors, such as origin and genetic inheritance, as well as environmental aspects such as adaptability to the planting environment, centralized management of breeders and extensive cultivation, which have led to increased gene exchange among the Zoysia populations[45]. Liu et al.[46] conducted genetic diversity analysis of 42 seashore paspalum (Paspalum vaginatum Sw.) based on morphology and SRAP molecular markers, and the results of the molecular marker clustering showed that germplasms from the same region were more likely to be clustered together, while the morphology clustering showed that the germplasms clustered together often originated from distinct geographical locations.
DNA fingerprints generated using molecular markers can offer more precise genetic information[47]. Molecular markers have been utilized to construct fingerprints to identify plant varieties, including radish and cucumber[48, 49]. Accurate identification of cultivars is essential for cultivation and breeding[50]. This study demonstrates that both SSR and SRAP are effective methods for constructing fingerprints of Zoysia germplasm. The 21 polymorphic bands amplified by one pair of SSR primer Xgwm234-5B were able to distinguish all 45 germplasm collections, while the SRAP molecular marker required a total of 43 bands amplified by two pairs of primers Me3-Em1 and Me3-Em2 to generate the fingerprints of the 45 germplasm collections. As Zoysia breeding advances, the number of varieties will continue to increase. To ensure the accuracy and reliability of the fingerprint of increasing number of new varieties, additional primers need to be developed, and it is essential to expand the fingerprint database of Zoysia. In this study, 45 Zoysia lines were divided into six clusters based on both SSR and SRAP markers and fingerprints of those plants were constructed. The findings of this study provided a theoretical basis for the subsequent evaluation and identification of Zoysia germplasm as well as new variety development.
This research was financially supported by the Fundamental Research Funds for the Central Universities (XUEKEN2022020).
-
The authors declare that they have no conflict of interest.
- Copyright: © 2023 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Wang M, Chen Q, Yu J, Liu J, Tate TM, et al. 2023. Genetic diversity analysis and fingerprint construction for 45 Chinese Zoysia germplasm collections. Grass Research 3:10 doi: 10.48130/GR-2023-0010
Genetic diversity analysis and fingerprint construction for 45 Chinese Zoysia germplasm collections
- Received: 15 March 2023
- Accepted: 20 April 2023
- Published online: 09 June 2023
Abstract: Zoysia spp. germplasm exhibit genetic variation between and within species. A comprehension of the genetic diversity of Zoysia germplasm could enable the effective utilization of these germplasms in future breeding endeavors. Ten simple sequence repeats (SSR) primer pairs and nine sequence-related amplified polymorphism (SRAP) primer pairs were used to analyze genetic diversity and construct DNA fingerprints for 45 Chinese Zoysia germplasm collections. We detected 231 SSR polymorphic bands and 149 SRAP polymorphic bands with 97.18% and 93.43% polymorphism ratios, respectively. The genetic similarity coefficient of the 45 germplasm collections ranged from 0.623 to 0.856, with an average of 0.727. Forty-five germplasm collections were divided into six major clusters when the genetic similarity coefficient was 0.71 based on the unweighted pair group method with the arithmetic averaging (UPGMA) method. Both SSR and SRAP molecular marker systems can be used to identify all germplasm collections, the SSR primer pair (Xgwm234-5B) and SRAP primer pairs (Me3-Em1 and Me3-Em2) can effectively distinguish 45 Zoysia spp. accessions. Collectively, we utilized both SSR and SRAP molecular markers to generate DNA fingerprints in this study providing a theoretical foundation for germplasm conservation and assisting in selecting and breeding new varieties of Zoysia.
-
Key words:
- Fingerprint /
- Genetic diversity /
- Zoysia spp. germplasm /
- SSR /
- SRAP












