-
Edible bird nest (EBN) has a long history of consumption in various cultures, particularly in East Asia. It is primarily derived from the nests of certain swiftlet species, such as the Edible-nest Swiftlet Aerodramus fuciphagus (Thunberg, 1812) or the Black-nest Swiftlet Aerodramus maximus (Hume, 1878). These nests are harvested for culinary use on a large scale. EBNs are valued in particular for their nutritional value, flavor and health benefit. EBNs are rare products and represent a delicacy, sometimes referred to as 'Caviar of the East', and with prices reaching up to several thousand USD/kg, they belong to the most expensive animal products used for nutritional purposes[1]. Allegedly, the consumption of EBN has been associated with numerous health benefits; it is claimed that EBNs are a good source of high-quality proteins, containing essential amino acids that are necessary for the growth and repair of body tissues[2]. They are also a source of essential minerals such as calcium, iron, potassium, and magnesium, which play crucial roles in maintaining proper bodily functions. Moreover, EBNs are known for their unique composition of complex carbohydrates which are covalently linked to their proteins, consisting mainly of N-acetylgalactosaminyl-, galactosyl-, N-acetylglucosaminyl- and N-acetylneuraminyl- moieties, providing the gelation and water holding properties of this functional food (Fig. 1). Arguably, the high concentration of these complex carbohydrates in EBN possesses immunomodulatory properties, which may help strengthen the body's immune system and enhance its ability to fight against diseases[3].
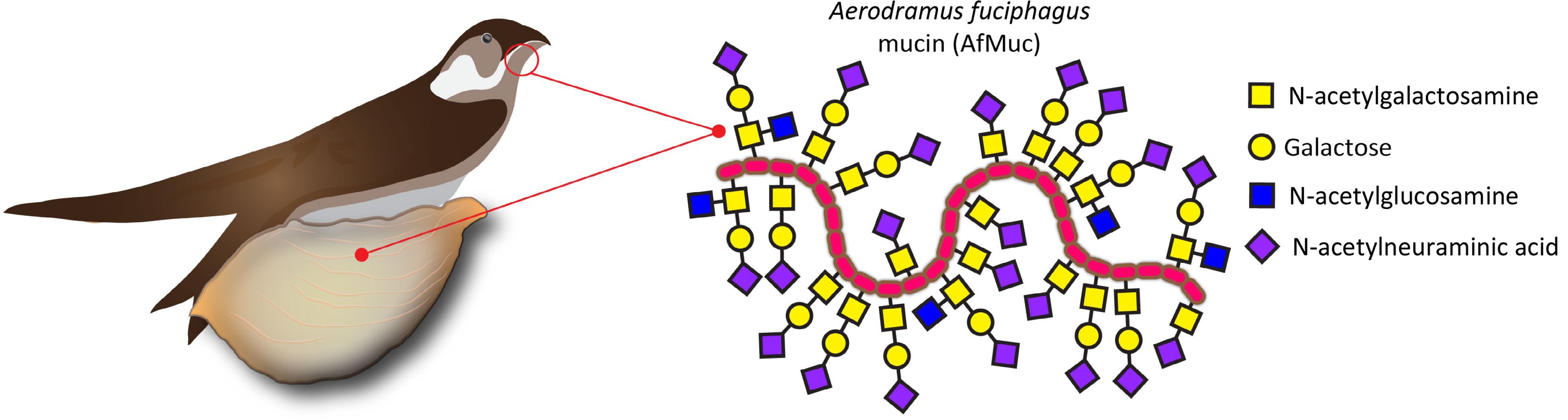
Figure 1.
EBN is produced by male swiftlets' salivary glands. The largest portion of this dried saliva consists of mucins, which are proteins heavily decorated with complex carbohydrates linked via N-acetylgalactosamine to the AfMuc protein backbone.
In recent years, the economic potential of EBN has gained attention. Although the current market volume is huge, reaching annual sales of several thousand tons per year, EBN is only harvested in certain regions of southeast Asia[4]. Due to the natural scarcity of EBN, the biotechnological production of EBN components is of considerable interest. The production of functional EBN components by biotechnological means for human nutrition would be valuable progress and would pave the way to a fully synthetic EBN.
Understanding the functional mechanisms and significance of protein O-glycosylation in EBN holds great potential for further exploration and exploitation of its health benefits. By unravelling the intricate relationship between protein O-glycosylation and the bioactive properties of EBNs, one can expect novel approaches for enhancing the production, quality, and bioactivity of EBN products, which can be provided at a significantly lower price than harvested EBN.
Herein, we describe the molecular basis of the initial glycosylation step of a previously unstudied mucin from Aerodramus fuciphagus (AfMuc) using various polypeptide N-acetylgalactosaminyltransferase isoforms. In addition, we biochemically compare a novel transferase enzyme originating from Aerodramus fuciphagus with an isoform from Homo sapiens (AfGalNT2 and HsGalNT2, respectively).
-
Sequencing read SRR4116906 derived from A. fuciphagus salivary gland de novo assembled transcripts deposited by Looi et al.[5] in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) was uploaded to the Galaxy web platform for DNA trimming and sequence assembly at usegalaxy.org[6]. The assembled protein sequences (Supplemental Figs S1−S10) were then searched with a local BLAST server[7] against human mucin-5AC and human polypeptide N-acetylgalactosaminyltransferase 2 (UniProt accessions P98088 and Q10471, respectively).
Materials
-
Escherichia coli Mach1 T1 cells (Life Technologies, Beijing, China) were used for plasmid cloning and maintenance, and strain E. coli BL21 (DE3) strain (Invitrogen, Shanghai, China) was used for expression experiments. The gene sequences for AfMuc, HsGalNT2, AfGalNT2, and CjCgTB30 (Supplemental Figs S1−S4, respectively) were synthesized in codon-optimized form for expression in E. coli K12 by Genscript Ltd. (Nanjing, China). Taq DNA polymerase, restriction endonucleases and T4 ligases were purchased from Thermo Fisher Scientific (Shanghai, China); DNA gel purification and plasmid extraction kits were purchased from Axygen (Beijing, China); Peptide substrates were synthesized from Changzhou Kanglong Biotech Ltd. (Shanghai, China). Nucleotide sugars were purchased from Carbosynth Ltd (Suzhou, China). All other chemicals used in this study were of the highest grade available from various major Chinese chemical distributors.
Recombinant protein expression and purification
-
The expression of recombinant enzymes was carried out as described previously[8]. Briefly, the pet30a expression plasmids containing the target gene sequences of AfMuc, HsGalNT2, AfGalNT2, and CjCgTB30 were transformed into the E. coli BL21 (DE3) expression host cells and grown on Luria Bertani (LB) agar plates supplemented with 50 μg/mL kanamycin. Singled-out colonies were picked and shaken overnight in 2 mL of LB medium containing 50 μg/mL kanamycin at 37 °C. Cells were then transferred into 400 mL of fresh LB medium and shaken at 37 °C with shaking at 250 rpm until the cell density reached an optical density of 0.5. The cultivation temperature was then reduced to 16 °C, and protein expression was induced by adding a final concentration of 1 mM of isopropyl β-D-thiogalactopyranoside (IPTG) to the cell suspension. After 24 h, cells were harvested by centrifugation (4,000 g for 15 min) and resuspended in 10 mL of cell lysis buffer (100 mM NaCl, 50 mM Tris, 1% Triton X-100 (v/v), adjusted with HCl to pH 8.0), and disintegrated by ultrasound sonication. After 20 min of centrifugation at 12000 g, the resulting supernatant was collected for further purification. The cleared supernatants were then loaded onto nickel-nitrilotriacetate (Ni-NTA) agarose affinity columns (3 mL bed volume, Qiagen, Beijing, China). The Ni-NTA columns were washed with 10 column volumes of washing buffer (50 mM Tris-HCl, 300 mM NaCl, 20 mM imidazole, pH 8.0). The desired proteins were eluted with elution buffer (500 mM imidazole, 50 mM Tris-HCl, 300 mM NaCl, pH 8.0) and imidazole and other buffer components were removed by replacing the buffer with water using centrifugal concentrators (10 kDa molecular weight cut-off, Sartorius, Germany). Protein bands were visualized using Coomassie brilliant blue G-250 staining. The purified and most active sample fractions were pooled and stored at −80 °C for further experiments.
In vitro glycosylation assays
-
The enzymatic activities of recombinant HsGalNT2 and AfGalNT2 were tested with chemically synthesized peptide donor substrates Muc1 (APGSTAPPA) and EA2 (PTTDSTTPAPTTR) using the glycosylation assays previously described[9,10]. Briefly, 10 µL mixtures consisting of 1 mM peptide substrate, 1 mM UDP-N-acetylgalactosamine (UDP-GalNAc), 1 mM MnCl2, 50 mM 2-(N-morpholino) ethane sulfonic acid (MES) buffer, pH 7.0, and ~12 µg of purified enzyme were incubated at 37 °C for 16 h. The glycosylated product was then further used for a galactosylation reaction using the recombinant β1,3-galactosyltransferase derived from Campylobacter jejuni (CjCgTB30) as follows: 10 µL mixtures consisting of 1 mM GalNAc-Muc1 glycopeptide substrate, 1 mM UDP-galactose (UDP-Gal), 1 mM MnCl2, 50 mM Tris/HCl buffer, pH 7.5, and ~2 µg of CjCgTB30 enzyme were incubated at 37 °C for 12 h. In some glycosylation experiments, UDP-GalNAc was replaced with UDP-GlcNAc.
Alternatively, glycosylation assays were also performed by substituting the peptide substrates with recombinant AfMuc protein backbone. The glycosylation reaction was performed in 20 µL mixtures consisting of ~1 mg/mL AfMuc protein backbone, 1 mM UDP-GalNAc, 1 mM MnCl2, 50 mM MES buffer, pH 7.0, and ~24 µg of purified enzyme were incubated at 37 °C for 16 h. All samples were subsequently analyzed by Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-ToF MS) analysis.
Biochemical characterization of recombinant HsGalNT2 and AfGalNT2
-
The pH optima of these enzymes were determined using sodium phosphate buffers (final concentration 100 mM) adjusted to various pH values in the range of pH 5.0−9.0. The temperature optima of HsGalNT2 and AfGalNT2 were determined by incubating the reaction mixtures at different temperatures ranging from 4 to 55 °C. The effect of metal ions (Ca2+, Co2+, Cu2+, Mg2+, Mn2+, Zi2+, Zn2+) or EDTA on the activity of the enzymes was tested at a final concentration of 1 mM.
MALDI-ToF MS analysis
-
MALDI-ToF MS was performed directly from reaction mixtures using a Bruker Autoflex Speed instrument (equipped with a 1,000 Hz Smartbeam-II laser) using 6-aza-2-thiothymine as a crystallization matrix. The obtained mass spectra were analyzed by Bruker Flexanalysis software version 3.3.80.
-
Most reported attempts at preparing recombinant HsGalNT2 were achieved using eukaryotic expression systems[11−15], but several recent works also described the soluble expression of this human enzyme in Escherichia coli[16−18]. Motivated by these findings, we also attempted the recombinant expression of HsGalNT2 and AfGalNT2 but found that most of the proteins were produced in an insoluble form (Supplemental Fig. S5). Nevertheless, Western blot analysis confirmed that a small portion of the recombinant proteins were also expressed in soluble form (Supplemental Fig. S6). Both purified HsGalNT2 and AfGalNT2 enzymes were able to glycosylate the glycopeptide substrates Muc1 (Fig. 2a−c, Supplemental Fig. S7) and EA2 (Supplemental Figs S8 & S9) using UDP-GalNAc or UDP-GlcNAc as sugar donors.
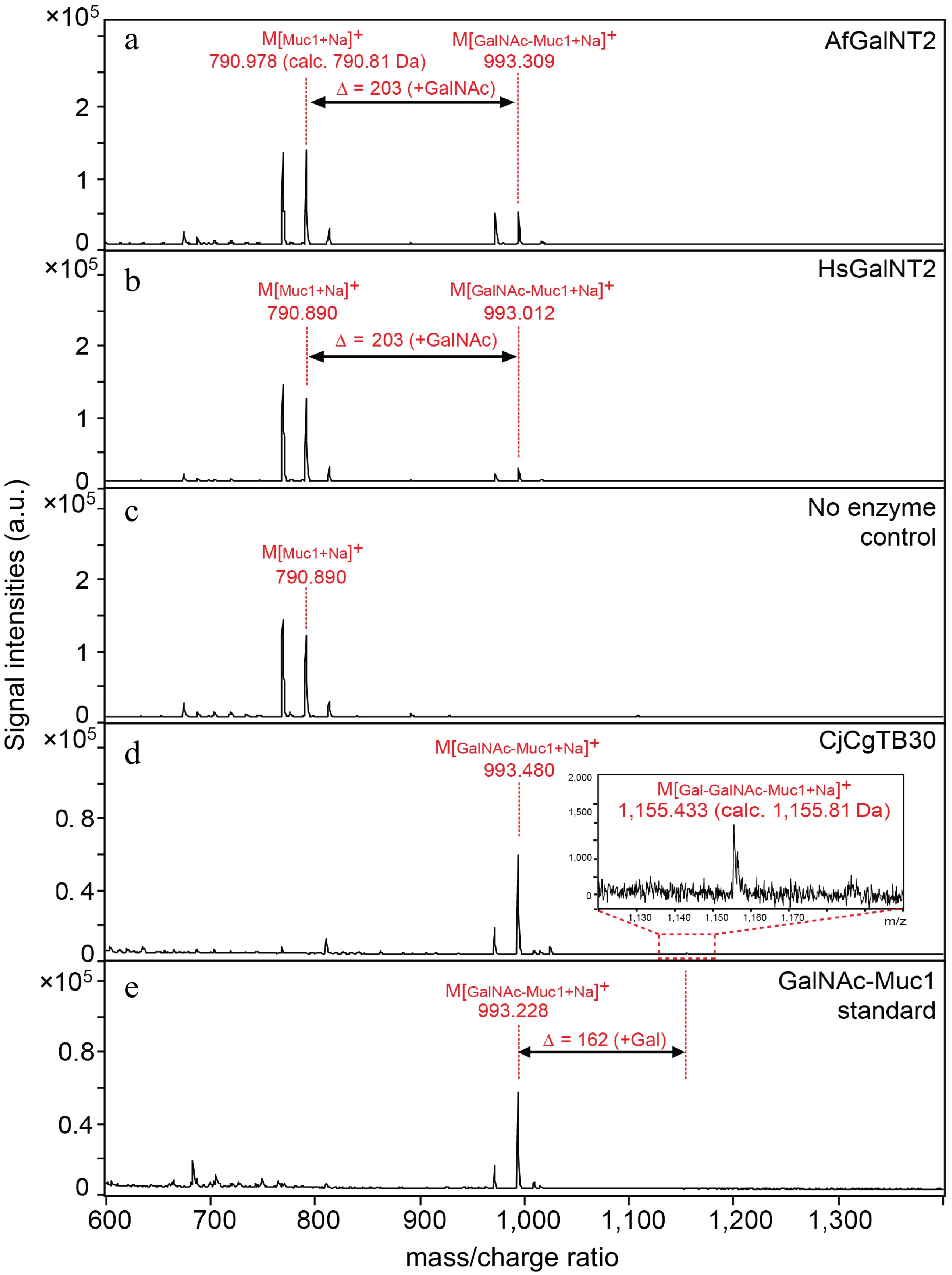
Figure 2.
Activity of AfGalNT2 and HsGalNT2 towards the Muc1 peptide substrate. Reaction mixtures containing the Muc1 peptide in the presence of UDP-GalNAc and (a) AfGalNT2; (b) HsGalNT2; (c) no enzyme; (d) Galactosylation reaction of the GalNAc-Muc1 glycopeptide substrate in the presence of CjCgTB30 enzyme and UDP-galactose; (e) reaction mixtures without CjCgTB30.
Du et al. recently reported the galactosylation of glycoproteins using a whole-cell bacterial expression platform by introducing a β1,3-galactosyltransferase derived from Campylobacter jejuni (CjCgTB30) into the expression host[16]. We therefore also used the same recombinant biocatalyst in purified form (Supplemental Fig. S10) to further galactosylate the GalNAc-Muc1 glycopeptide. Although the reaction product could be clearly identified by MALDI-ToF MS (Fig. 2d−e), the conversion rates of this galactosylation reaction were rather low (< 10%). However, given the importance of the GalNAc-β1,3-Gal moiety in various areas of medical and bioanalytical sciences[19−21], the ability to produce even small (but chemically well-defined) amounts of this motif is highly desirable.
Biochemical characterization of HsGalNT2 and AfGalNT2
-
The pH optima of AfGalNT2 and HsGalNT2 were evaluated in a pH range of 5.0−9.0 (Fig. 3a). Both enzymes showed comparable pH dependencies with maximal activities at pH 7.0. These pH optima are in the expected range for enzymes, as previously described polypeptide N-acetylgalactosamine transferase isoforms reported pH optima between 6.0 and 8.2[10,22,23].
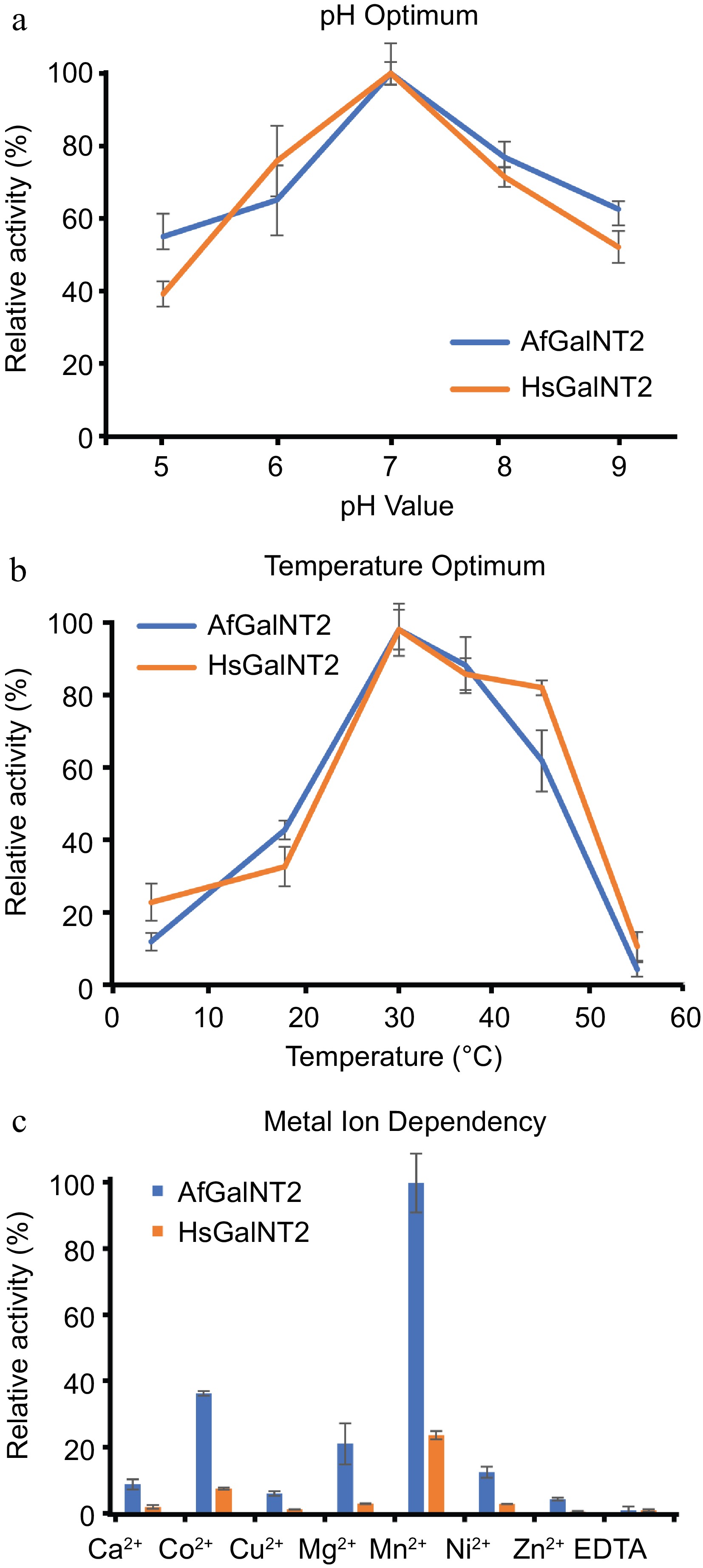
Figure 3.
Biochemical characterization of AfGalNT2 and HsGalNT2. The following parameters were measured for both enzymes using the EA2 peptide: (a) pH optimum, (b) temperature optimum and (c) metal ion dependence.
Testing AfGalNT2 and HsGalNT2 at various temperatures (4, 18, 30, 37, 45, and 50 °C) at pH 7.0 with the optimized conditions described above showed that both enzymes had their maximum activity at 30 °C (Fig. 3b). These values are lower than the temperature optima of 42 °C described for isoforms 6 and 16 from Xenopus laevis[10], but higher than the optimal temperatures of 23 and 28 °C reported for isoforms from Caenorhabditis elegans[24] and Drosophila melanogaster[25].
Previous studies described divalent manganese ions as the best activator for glycosylation reactions of polypeptide N-acetylgalactosaminyl transferases[22,26]. Figure 3c shows the screening of metal dependencies of the enzymes when exposed to various divalent metal ions in their chloride form at a concentration of 1 mM, which resulted in comparable activities in the presence of various divalent cations, H2O and EDTA. The obtained data are in agreement with reported studies on previously studied polypeptide N-acetylgalactosaminyl transferases[10,27]. Mn2+-ions had the strongest effect on the enzymes' activities, whereas Co2+, Mg2+, Ca2+ and Cu2+ also stimulated the glycosylation reaction only to a smaller extent.
Expression, purification and in vitro glycosylation of AfMuc
-
So far, the recombinant expression of soluble mucin-protein backbones in bacterial expression systems was not described yet. Recent works which transferred GalNAc moieties onto protein donors used non-mucin-like protein backbones derived from interleukin-2[18] or interferon α-2b[16], and reported the addition of only a single GalNAc residue to the protein backbone using whole-cell biotransformation reactions in E. coli. As shown in Fig. 4a, AfMuc could be successfully expressed and purified to near homogeneity. Mass spectrometric analysis of the whole protein showed good agreement between the measured molecular mass and theoretical (calculated) mass (18,069.905 Da, calc. 18,071 Da, Fig 4b). Furthermore, glycosylation reactions in the presence of HsGalNT2 and UDP-GalNAc showed apparent and discrete increases of the m/z values by ~203 Da, indicating the addition of up to four GalNAc residues (Fig. 4c).
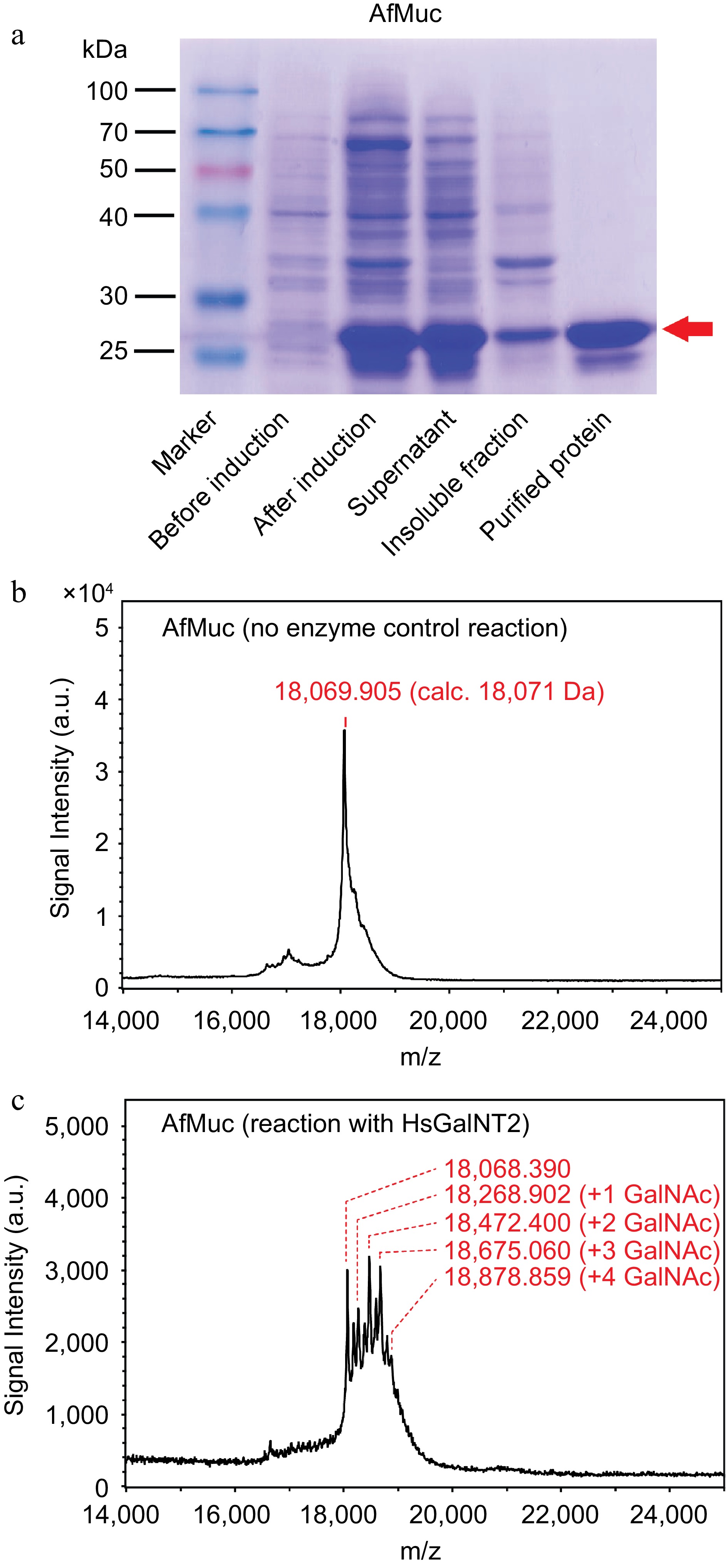
Figure 4.
Purification and in vitro glycosylation of AfMuc. (a) SDS-PAGE analysis of AfMuc during expression and purification. The red arrow indicates the position of the desired purified protein. (b) MALDI-ToF MS analysis of recombinant AfMuc without the presence of HsGalNT2. (c) MALDI-ToF MS analysis of recombinant AfMuc in the presence of HsGalNT2. The intermediate-mass peaks between discrete GalNAc units may be the result of terminal proteolysis of the recombinant AfMuc protein backbone.
-
This work described the identification, bacterial expression, purification and in vitro glycosylation of a mucin protein (AfMuc) derived from the Edible-nest Swiftlet Aerodramus fuciphagus. In addition, we biochemically characterized an unstudied polypeptide N-acetylgalactosaminyltransferase from the same swiftlet species in recombinant form and compared it with a related human enzyme. The herein-presented enzymes and methods may be of use for further exploration and exploitation of the functional components of EBN using biotechnological production methods. However, as for all recombinant proteins expressed in bacteria which are used as food additives to enhance various aspects of food processing and quality, there is a potential risk of contaminants associated that need to be considered. These contaminants can include bacterial components, such as lipopolysaccharides, peptidoglycans, nucleic acids, and other cellular components. In addition to residual growth media components, such as sugars, amino acids, and vitamins, also chemical residues from the addition of agents like antibiotics, detergents, or solvents may persist. Given that, this work conceptualized the overall feasibility of the in vitro glycosylation of EBN mucin only in a laboratory setup, further improvements to reduce costs, possible contaminants and increase the overall expression titers and glycosylation efficiency during the large-scale production of EBN will require biotechnological and process engineering efforts.
-
The authors confirm contribution to the paper as follows: study conception and design: Liu L, Voglmeir J; data collection: Cheng G, Lyu Y, Ran R; analysis and interpretation of results: Liu L, Voglmeir J; draft manuscript preparation: Liu L, Voglmeir J. All authors reviewed the results and approved the final version of the manuscript.
-
To be supplied by author.
-
The authors declare that they have no conflict of interest.
- Supplemental Fig. S1 Gene sequence of the AfMuc gene with the underlined NdeI/XhoI restriction sites. The N-terminal octa-histidine tag is shown italicized letters. AfMuc was subsequently cloned into the pET30a expression vector.
- Supplemental Fig. S2 Gene sequence of the HsGalNT2 gene with the underlined NdeI/XhoI restriction sites. HsGalNT2 was subsequently cloned into the pET30a expression vector.
- Supplemental Fig. S3 Gene sequence of the AfGalNT2 gene with the underlined NdeI/XhoI restriction sites. AfGalNT2 was subsequently cloned into the pET30a expression vector.
- Supplemental Fig. S4 Gene sequence of the CjCgtB30 gene with the underlined NdeI/XhoI restriction sites. CjCgtB30 was subsequently cloned into the pET30a expression vector.
- Supplemental Fig. S5 SDS-PAGE analysis of HsGalNT2 (top) and AfGalNT2 (bottom) during expression and purification. The red arrows indicate the expected molecular weights of the purified recombinant proteins.
- Supplemental Fig. S6 Western-blot analysis of the puridied protein fractions of HsGalNT2 (top) and AfGalNT2 (bottom). The Western-blot analysis was performed by transferring proteins from the protein gels to nitrocellulose using a semi-dry blotting apparatus. After blocking with 0.5% BSA, the membranes were incubated with horseradish peroxidase-conjugated mouse anti-polyHistidine antibody (anti-His, 1:1000 dilution, Genscript, Nanjing); after washing, recombinant protein was visualized chromogenically using 3,3-diaminobenzidine reagent.
- Supplemental Fig. S7 Activity of AfGalNT2 and HsGalNT2 towards the Muc1 peptide substrate Reaction mixtures containing the Muc1 peptide in the presence of UDP-GlcNAc and (A) AfGalNT2; (B) HsGalNT2; (C) no enzyme control.
- Supplemental Fig. S8 Activity of AfGalNT2 and HsGalNT2 towards the EA2 peptide substrate Reaction mixtures containing the EA2 peptide in the presence of UDP-GalNAc and (A) AfGalNT2; (B) HsGalNT2; (C) no enzyme control.
- S9
- Supplemental Fig. S9 Activity of AfGalNT2 and HsGalNT2 towards the EA2 peptide substrate Reaction mixtures containing the EA2 peptide in the presence of UDP-GlcNAc and (A) AfGalNT2; (B) HsGalNT2; (C) no enzyme control.
- Supplemental Fig. S10 SDS-PAGE analysis of the recombinant β1,3galactosyltransferase derived from Campylobacter jejuni (CjCgTB30) during expression and purification. The red arrow indicates the expected molecular weight of the purified recombinant protein.
- Copyright: © 2024 by the author(s). Published by Maximum Academic Press on behalf of Nanjing Agricultural University. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Cheng G, Lyu Y, Ran R, Liu L, Voglmeir J. 2024. Expression and in vitro glycosylation of recombinant edible bird nest (EBN) mucin. Food Materials Research 4: e002 doi: 10.48130/fmr-0023-0037
Expression and in vitro glycosylation of recombinant edible bird nest (EBN) mucin
- Received: 13 July 2023
- Revised: 22 August 2023
- Accepted: 06 November 2023
- Published online: 02 January 2024
Abstract: Edible bird's nest (EBN) is considered a delicacy in Chinese cuisine. The nests are produced by male swiftlets (Aerodramus sp.). Due to their high price, the nutritional benefit from EBN can currently only be afforded by affluent consumers. Therefore, the ability to identify and produce recombinant, functional EBN mucins is highly desirable. Herein, we report the identification, bacterial expression, purification and glycosylation of the first recombinant mucin derived from the Edible-nest Swiftlet Aerodramus fuciphagus (AfMuc). Recombinant AfMuc was successfully expressed at high expression levels and purified to near homogeneity by nickel-chelation affinity chromatography. The in vitro glycosylation of the recombinant AfMuc protein backbone resulted in the addition of up to four GalNAc residues and could be achieved by using a human and a previously unstudied Aerodramus fuciphagus polypeptide N-acetylgalactosaminyltransferase (HsGalNT2 and AfGalNT2, respectively). A comparison of the biochemical properties of these enzymes revealed characteristics of these enzymes.













