-
Lignin, a complex and versatile polymer derived from lignocellulosic biomass, is one of the most prevalent biopolymers on earth that serve as a huge high-quality carbon sink. Lignin has a higher carbon content than cell wall polysaccharides such as cellulose and the hemicelluloses. The polyphenolic characteristics of lignin provide plants mechanical strength, hydrophobicity, and resistance to pathogens. However, lignin drastically hampers the processing and utilization of plant biomass. For example, there were negative correlations between lignin content in plant biomass and pulp yield, forage digestibility, or polysaccharide saccharification yield[1−3].
The backbone of lignin polymers is made up of three basic building blocks including p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) lignin units which are derived from three p-hydroxycinnamyl alcohols (termed as 'monolignols'), p-coumaryl, coniferyl, and sinapyl alcohols, respectively (Fig. 1a). The biosynthesis of traditional monolignols via the phenylpropanoid pathway is initiated by the conversion of the aromatic amino acid phenylalanine or tyrosine to cinnamic or p-coumaric acids via the phenylalanine ammonia lyase (PAL) or tyrosine ammonia lyase (TAL), respectively. The carboxylic acids are sequentially reduced to CoA-thioester, aldehyde, and alcohol by 4-coumarate coenzyme A ligase (4CL), cinnamoyl-CoA reductase (CCR), and cinnamyl alcohol dehydrogenase (CAD). The aromatic ring in cinnamate is hydroxylated via cinnamate 4-hydroxylase (C4H), a 'shikimate shunt' including p-hydroxycinnamoyl-CoA: quinate/shikimate hydroxycinnamoyl transferase (HCT), p-coumaroyl shikimate 3′ -hydroxylase (C3′H), and caffeoyl shikimate esterase (CSE), p-coumarate 3-hydroxylase/ascorbate peroxidase (C3H/APX), and ferulate 5-hydroxylase (F5H), then is 3-O-methylated and 5-O-methylated via caffeoyl-CoA O-methyltransferase (CCoAOMT) and caffeic acid O-methyltransferase (COMT), eventually yielding the three monolignols[4,5].
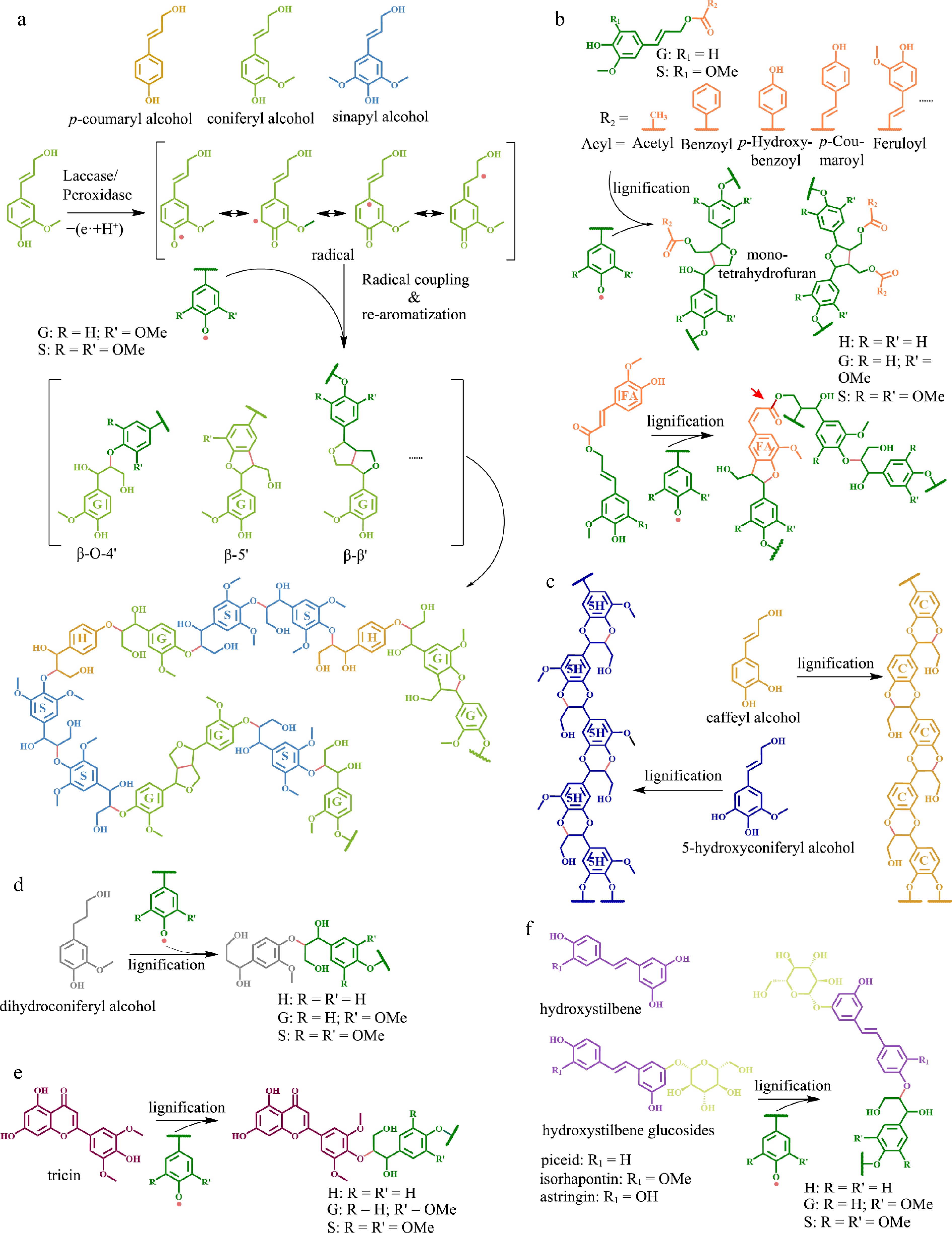
Figure 1.
Different types of monomers and corresponding lignin structures. (a) Conventional monolignols and their polymerization mechanism, which is mainly carried out by an 'end-wise' process that begins with single-electron oxidation of monolignols by laccase and/or peroxidase to generate activated radical intermediates. The radicals inter-couple or couple with a radical formed on the free-phenolic ends of a growing lignin polymer to form lignin with different linkages. (b) Acylated monolignols and the resulting lignin structure. Feruloyl esters, highlighted in orange color on feruloyl monomers, can be integrated into the lignin backbone. The red arrow indicate the presence of an alkali-labile linkage in the lignin structure. (c) Caffeyl alcohol and 5-hydroxyconiferyl alcohol (unconventional lignin monomers) and the resulting homogeneous linear lignin polymer. (d) Dihydroconiferyl alcohol and the corresponding lignin structure. (e) Tricin (a flavonoid) and the resulting lignin polymer. (f) Hydroxystilbene and hydroxystilbene glucoside, as well as their corresponding lignin structures.
In addition to traditional monolignols, researchers have recently identified several essential unconventional phenolic compounds that can also serve as monomers participating in oxidative radical reactions and integrating into lignin polymers in certain plants. These unconventional monomers included phenolic compounds emerging from typical monolignol biosynthesis pathways (e.g., acylated monolignols, caffeyl and 5-hydroxyconiferyl alcohols, and hydroxycinnamaldehydes) or other phenolic biosynthesis pathways, including flavonoids (e.g., tricin and naringenin), hydroxystilbenes (e.g., resveratrol, piceatannol, and isorhapontigenin) and their O-glucosylated counterparts (e.g., piceid, astringin, and isorhapontin), and ferulic acid amides (e.g., feruloyltyramine, and diferuloylputrescine)[6]. The identification of these unconventional monomers demonstrated that lignin allows compositional flexibility and structural plasticity[6,7], opening up new avenues for engineering the lignin structure.
Monolignols are transported into the cell wall in a manner that is not fully comprehended[8]. Polymerization occurs via free-radical coupling mediated by peroxidases and/or laccases, generating various structures in the biopolymers[4,9,10] (Fig. 1a). Structural characteristics of lignin are primarily determined by types of inter-unit linkages and functional groups attached to the monomers. There are two major types of inter-unit linkages present in lignin polymers, including ether bond linkages (e.g., β-O-4 and 4-O-5) and carbon-carbon linkages (e.g., β-β, β-5, 5-5, and β-1). The inter-unit linkages in natural lignin exhibit variation across plant species, with the β-O-4 linkage being the most prominent[11]. The main functional groups present in lignin include hydroxyl, methoxyl, carbonyl, carboxyl, and benzene ring groups. The proportion of each group in lignin highly depends on the types of monomers and the method of lignin processing[12].
The molecular structures of lignin, particularly the composition, inter-unit linkages, molecular weight, and solubility have a significant impact on the physicochemical properties of lignin and the usability of lignocellulosic biomass. Unconventional monomers expand the definition of traditional lignin and provide new strategies for tailoring the structure of lignin. Recently, several review reports have discussed the occurrence and biosynthesis of unconventional monomers, as well as the structural features of unconventional lignin[6,7,13]. Here, we mainly focus on the potential utilization of those unconventional monomers, review the progress of unconventional lignin engineering, and anticipate future strategies.
-
The lignin in a variety of plant taxa is decorated with acylesters, including p-coumarate (pCA), ferulate (FA), p-hydroxybenzoate (pBA), benzoate (BA), vanillate (VA), and acetate[14]. The pCA decoration is one of the hallmarks of the commelinid monocots[15,16] and has also been found in several eudicot species such as mulberry (Morus alba) and kenaf (Hibiscus cannabinus)[17,18]. The pBA decoration was reported in palms, poplars, and willows[19−22], as well as in seagrass Posidonia oceanica[23,24]. The acetate decoration at low levels was ubiquitous among angiosperms with substantial levels in the lignin of kenaf (Hibiscus cannabinus), sisal (Agave sisalana), abaca (Musa textilis), and hornbeam (Carpinus betulus)[7,25]. The FA decoration was also found in various angiosperms[26]. The BA decoration was reported in the lignin of some plants, including the macaúba (Acrocomia aculeata) palm fruit endocarp[22] and the PtrC4H1/PtrC4H2/PtrC3H3 transgenic lines of Populus trichocarpa[27]. Lignin from the leaves of Canary Island date palms was decorated with pCA, FA, pBA, BA, VA, and acetate[28]. The pCA, pBA, and acetate were preferentially decorated on syringyl units[14,29,30], whereas FA preferentially acylated guaiacyl units in eudicot lignin and syringyl units in monocot lignin[26].
Monolignols undergo acylation prior to being incorporated into the lignin polymer[14,31]. The acylated monolignols or monolignol γ-carboxylate conjugates behave as authentic lignin monomer precursors incorporated into the lignin polymer by polymerization and co-polymerization with the traditional monolignols. These monolignol ester conjugates can form identical linkages as traditional monolignols except β-β which is greatly influenced by the acylation (Fig. 1b)[13]. The inter-coupling of monolignols or coupling to growing polymer produces a quinone methide intermediate, which is rearomatized via nucleophilic attack to form various structures (Fig. 1a). Contrary to traditional monolignols β-β coupling, which involves rearomatization of intermediate quinone methides via free γ-OH groups attack to form a resinol structure, acylated monolignols preclude rearomatization via intramolecular cyclization and form a mono-tetrahydrofuran structure (Fig. 1b)[13]. Acylated monolignols may increase lignin polymer alkaline solubility by introducing alkaline labile ester bonds into lignin. However, different acylated monolignols exhibit distinct impacts on lignin properties. For instance, monolignol p-coumarate conjugate and monolignol p-hydroxybenzoate conjugate introduced an additional free phenolic 4'-hydroxyl group into lignin polymer in contrast to monolignol benzoate and monolignol acetate (Fig. 1b). Therefore, acylated lignin possessing different monolignol conjugates may exhibit distinct hydrophobicity since phenolic hydroxyl can increase lignin hydrophobicity[32]. Incorporation of monolignol ferulate into the backbone of lignin polymer via both monolignol and ferulate moieties (Fig. 1b) can produce more branched lignin[13,33]. However, some other acylated monolignols, such as monolignol p-coumarate and monolignol p-hydroxybenzoate, can only be integrated into the lignin polymer via their monolignol moieties. Therefore, these ester-linked phenolics can be found pendant on the lignin polymer[14].
Monolignol acylation requires acyltransferases to conjugate the activated acyl moieties from the thioester donor to traditional monolignols. The enzymes identified for p-coumaroylation of monolignols in grasses included p-coumaroyl CoA: monolignol p-coumaroyltransferase (PMT) from Oryza sativa (OsPMT)[34], Branchypodium distachyon (BdPMT)[35] , Zea mays (ZmpCAT)[36], and an eudicot species Hibiscus cannabinus (HcPMT)[18]. The enzyme reported for feruloylation of monolignols in both monocots and eudicots included feruloyl CoA:monolignol feruloyltransferase (FMT) from Angelina sinensis (AsFMT)[33] and O. sativa (OsFMT1)[26]. Recently, an enzyme p-hydroxylbenzoyl CoA: p-hydroxylbenzoyltransferase (PHBMT1) was characterized for p-hydroxylbenzoylation of monolignols in poplar[37,38]. Consistent with the discovery of lignin p-coumaroylation and p-hydroxylbenzoylation primarily on syringyl units, the recombinant OsPMT, ZmpCAT, HcPMT and PHBMT1 preferentially acylated sinapyl alcohol in vitro[18,34,36,37]. Contrarily, the recombinant AsFMT acylated all three monolignols with a preference for coniferyl alcohol[33]. So far, all identified enzymes catalyzing monolignol acylation belong to the well-known superfamily of plant BAHD acyltransferases. It is believed that additional BAHD acyltransferases, as well as the enzymes responsible for substrate supply, are also involved in the biosynthesis of acylated monolignols.
Although it is unclear how structural changes in acylated lignin will impact its utilization, acylation has been shown to affect its physicochemical properties and biomass recalcitrance[23,33,37]. Thus, the utilization of acylated lignin holds significant potential in various applications, including lignin separation, lignocellulosic biomass saccharification, lignin valorization, and the development of durable carbon-capture biomass. For example, overexpression of AsFMT in Arabidopsis and hybrid poplar (Populus alba × grandidentata) resulted in 'Zip lignin' via the incorporation of monolignol FA conjugates into the lignin polymer backbone[33]. The chemically labile linkages in 'Zip lignin' increased the efficiency of lignin depolymerization and the digestibility of the cell wall biomass following a mild alkaline treatment[33,39]. Additionally, heterologous expression of PMT in Arabidopsis and poplar resulted in the incorporation of novel pCA groups into the lignin of transgenic plants[40,41]. Similarly, overexpression of PHBMT1 in poplars increased the pBA amount in the lignin of transgenic plants[37,38]. The pendent pCA and pBA groups are readily clipped off from lignin upon a mild alkaline treatment, therefore, they can serve as an alternate source for producing platform chemicals. Therefore, engineering acylated monolignols can be a promising strategy for lignin valorization.
The precise biological roles of acylated lignin as well as the biosynthetic pathway of those acylated monolignols are yet unclear, impeding the progress of acylated lignin engineering. A comprehensive understanding of the biosynthesis of acylated monolignols and identifying additional key enzymes engaged in these pathways may lead us to new strategies and tools to engineer lignin with enhanced lignocellulosic biomass utilization.
-
Heterogeneity has long been an issue restricting the separation and utilization of lignin. Classic lignin is a complex heteropolymer of two to three different monomers with a variety of inter-unit linkages that are highly dependent on the supply of the individual monolignols at the lignification sites. Two unconventional monomers caffeyl alcohol and 5-hydroxyconiferyl alcohol containing O-diphenol groups form benzodioxane structures after a rapid intramolecular trap of quinone methide intermediates during lignification to generate homogeneous and linear all-ether-linked C-lignin and 5HG-type lignin polymers, respectively (Fig. 1c)[42−44]. The aforementioned polymers have several desirable characteristics that are absent in traditional lignin, thereby rendering them suitable for effective utilization of lignin. Firstly, lignin polymers containing benzodioxane structures do not form condensed units under acidic industrial lignin fractionation[45], thus they are more stable than traditional lignin polymers, which makes lignin separation and utilization easier. Secondly, lignin homopolymer can only be degraded into a few products (a single monomer in a high proportion) via hydrogenolysis[46,47], providing a promising strategy for the valorization of lignin. Thirdly, the high-purity linear lignin itself can serve as a good raw material to produce value-added biomaterials such as high-performance carbon fibers. It is reported that C-lignin-based carbon fiber outperformed conventional Kraft lignin carbon fiber in terms of thermal stability, molecular weight, diameter, and crystallinity[48]. Additionally, the benzodioxane linkages in lignin confer distinctive thermodynamic properties during co-polymerization and the linear structure facilitates the formation of different types of crystals in the co-polymer, making C-lignin a promising candidate for the development of new polymeric materials[46]. Therefore, the unique characteristics of homogeneous linear lignin make it an 'ideal lignin' to facilitate lignocellulosic biomass utilization[45].
Natural C-lignin was reported mainly in the seed coats of some orchids, cacti, Euphorbiaceae and Cleome hassleriana[42,43,47,49,50] and 5HG type lignin was detected in the seed coats of three species from Escobaria[43]. Recently, multiple approaches have been or are being considered to achieve a substantial yield of C-lignin in biomass crops, including poplar and alfalfa[51].
Research on C-lignin biosynthesis in C. hassleriana has made great progress in providing potential targets and tools for C-lignin genetic engineering. C-lignin started accumulating in the seed coat of C. hassleriana about 12−14 d after pollination[49,52]. The accumulation of C-lignin in C. hassleriana was accompanied by changes in the expression profiles of genes involved in the monolignol biosynthesis pathway[50]. For instance, downregulation of CCoAOMT and/or COMT inhibited the methylation of the phenolic hydroxyl group in monolignol which was long known to increase the accumulation of caffeyl alcohol, a monolignol precursor for C-lignin[42,43]. Lignin from Pinus radiata ccoaomt mutant plants was reported to integrate a trace amount of caffeyl alcohol[53]. The proportion of C-lignin in 10-day-old comt ccoaomt double mutant Medicago truncatula seedlings was more than four-fold (~1.8% of total monolignol units) higher than that in wild-type plants, however, the absolute amount of lignin in the double mutant was greatly reduced[51]. Thus, these enzymes are the primary targets for engineering C-lignin in different plants.
The concurrent down-regulation of COMT and CCoAOMT in the hairy roots of M. truncatula increased C-lignin accumulation in transgenic lines by ~15% of total lignin[51]. However, downregulation of CCoAOMT and/or COMT in Arabidopsis, alfalfa, and poplar exhibited an inability to accumulate C-lignin, impaired growth, and decreased total lignin content[54−56], demonstrating that this strategy is not suitable for all plant species. The 5HG monomers were slightly increased in the stem of Arabidopsis comt mutant[56], indicating that inhibition of COMT activity is critical for 5HG-type lignin biosynthesis. Nevertheless, overexpression of F5H in an Arabidopsis comt line increased the amount of 5-hydroxylconiferyl alcohol in lignin at the expense of plant fitness and total lignin content[57].
Other key enzymes in the monolignol biosynthesis pathway that regulate caffeyl alcohol and 5-hydroxylconiferyl alcohol in planta may be potential targets for homogeneous linear lignin engineering. For example, the decreased expression of ChCAD4 and increased expression of ChCAD5 during C-lignin deposition in the seed coat of C. hassleriana indicated that ChCAD5 preferentially reduced caffealdehyde to caffeyl alcohol while ChCAD4 preferentially reduced coniferaldehyde to coniferyl alcohol[50]. Downregulation of ChCAD5 reduced the C/G ratio of lignin in the Cleome seed coat, suggesting that ChCAD5 was required for C-lignin biosynthesis[50]. Additionally, a seed coat-specific laccase from C. hassleriana, ChLAC8, exhibited the same temporal expression pattern as C-lignin accumulation and had a strong substrate preference for caffeyl alcohol[58]. The increase of C-unit upon expression of ChLAC8 in comt mutants of M. truncatula and Arabidopsis suggested that this enzyme promoted C-lignin formation in planta[58]. However, a systematic study on engineering C-lignin in M. truncatula hairy roots suggested that C-Lignin accumulation required strong down-regulation of both COMT and CCoAOMT instead of an expression of a heterologous LAC, CAD, or CCR with a preference for caffeyl alcohol[51].
Although it is promising to use genetic engineering for producing homogeneous linear polymers, existing strategies via interference of the key enzymes in the classic monolignol biosynthesis pathway result in low C-lignin accumulation, reduced lignin content, and stunted plant growth. Possible causes of these issues include a) an unstable supply of caffeyl alcohol and/or 5-hydroxylconiferyl alcohol precursor which is partially due to rapid conversion of the highly reactive O-diphenol into other metabolites; b) blockage of the O-methylation process which may prevent the normal development of certain tissues, such as vascular tissue; and c) the disruption of metabolism homeostasis in plants arising from intricate interplay between the lignin pathway and other primary and secondary metabolic pathways. Understanding these limitations is essential for rationally designing strategies for homogeneous lignin engineering. Additionally, AI-assisted directed evolution of enzymes can be a breakthrough for the production of novel and specific tool enzymes and advances in homogeneous lignin engineering.
-
Although there is no specific formula weight for lignin due to its heterogeneity, the average molecular weight of lignin can be estimated via several analytical techniques including GPC, MALDI-ToF MS, light scattering analysis, and NMR end-groups titration[59]. Additionally, the average degree of polymerization (DP) of lignin can be calculated by the monomeric composition and the relative abundance of lignin units[59]. Low molecular weight lignin is anticipated to be more easily extractable, leading to enhanced biomass processing. Moreover, the higher amount of free phenolics in lower molecular weight lignin can improve its antioxidant activity. For instance, the low molecular weight lignin showed stronger antioxidant activity when 15 different types of lignin were blended with polypropylene[60]. Another study evaluated 21 ethanol organosolv lignin samples as potential antioxidants and found that the lignin with more phenolic hydroxyl groups, fewer aliphatic hydroxyl groups, low molecular weight, and narrow polydispersity had high antioxidant activity[61].
Lignin monomers with fewer radical coupling sites may inhibit lignin elongation during active lignin polymerization, resulting in short lignin. Several unconventional monomers including hydroxybenzaldehydes from engineered Arabidopsis[62], dihydroconiferyl alcohol (Fig. 1d) and guaiacylpropane-1,3-diol in gymnosperms[63,64], as well as tricin (Fig. 1e) in grass[65] are capable of only a single coupling reaction via their hydroxyl group located on fourth position in the benzene ring, suggesting that they may only serve as lignin initiators. Incorporation of such monomers into lignin can reduce its length and molecular weight. For instance, expression of the bacterial hydroxycinnamoyl-CoA hydratase-lyase (HCHL) gene, which could cleave the propanoid side-chain of a hydroxycinnamoyl-CoA lignin precursor to generate the corresponding hydroxybenzaldehydes, in the lignifying tissues of Arabidopsis inflorescence stem resulted in the accumulation of side-chain-shorten monomer, decreased lignin chain length, and increased saccharification yield of biomass[62]. Engineering low molecular-weight lignin is a promising strategy to increase lignocellulosic biomass processing efficiency as well as valorizing lignin. Therefore, the identification of additional potential genes will make a worthwhile contribution to engineering low molecular-weight lignin in the future.
-
Diverse functional groups with varying polarity impart hydrophobic or hydrophilic characteristics to complex lignin polymer. Hydrophobic lignin is essential for transporting water and nutrients in the plant vascular system. However, the solubility of lignin is largely affected by its hydrophobicity. Increasing the hydrophilicity of lignin can render lignin more water soluble and reduce hydrophobic interactions between lignin and hemicelluloses, which may improve biomass saccharification yield[44]. Contrarily, increasing the hydrophobicity of lignin can inhibit the enzymatic hydrolysis of cellulose[66], thereby enhancing the durability of lignocellulosic biomass during storage.
The number of hydroxyl and carboxylic groups in lignin significantly affects its hydrophobicity. Unlike aliphatic hydroxyl, phenolic hydroxyl can increase lignin hydrophobicity[32]. The addition of certain unconventional monomers into lignin can significantly alter the quantity and type of hydroxyl groups in lignin polymer. For example, monomers substituted with sugar moieties such as hydroxystilbene glycosides (Fig. 1f) found in the lignin of Norway spruce bark increased the aliphatic hydroxyl group[67,68]. In contrast, the esterification of the aliphatic hydroxyl group at the γ-position of traditional monolignols, including monolignol p-hydroxybenzoate conjugate and monolignol p-coumarate conjugate, introduced an additional free phenolic 4'-hydroxyl group into lignin. Therefore, incorporating these acylated monomers into the lignin polymer can boost its hydrophobicity. Interestingly, it was reported that the higher quantity of monolignol p-hydroxybenzoate conjugate in seagrass Posidonia oceanica might be associated with the higher organic carbon storage capacity of its ecosystem[23].
Since the hydrophobicity of lignin is crucial for the normal growth of plants, future genetic engineering efforts to reduce the hydrophobicity of lignin must take full advantage of the natural unconventional monomers and focus on fine-tuning lignification in plants.
-
The artificially lignified maize cell walls system offered the potential to utilize a variety of phenolic compounds, including flavonoid derivatives, guaiacyl butenol, rosmarinic acid, caffeic acid derivatives, ferulic acid derivatives, coniferyl and sinapyl glucosides, and sinapoyl glucose as alternative monomers to form novel lignin in vitro[69−71]. Thus, it is conceivable to tailor the lignin structure in planta by substituting some fraction of the traditional monolignols with those alternative monomers through metabolic engineering to enhance lignocellulosic biomass utilization. Some chemically labile linkages can be introduced into lignin via alternative monomers, thereby significantly increasing the saccharification efficiency of engineered biomass. For instance, curcumin (diferuloylmethane) (Fig. 2a) was successfully incorporated into lignin polymer of Arabidopsis through simultaneous expression of Curcuma longa's Diketide-CoA synthase (DCS) and curcumin synthase 2 (CURS2) driven by the secondary cell wall CELLULOSE SYNTHASE A4 (CesA4) promoter[72]. The engineered plants showed normal growth and their lignocellulosic biomass incorporating curcumin into lignin had a 14%–24% higher saccharification efficiency following alkaline pretreatment as compared with the wild type[72]. Likewise, a coumarin compound called 'scopoletin' (Fig. 2b) was also incorporated into the lignin polymer of Arabidopsis via simultaneous expression of FERULOYL-CoA 6'-HYDROXYLASE 1 (F6'H1) and COUMARIN SYNTHASE (COSY) driven by the CesA4 promoter[73]. The transgenic events improved the saccharification efficiency of engineered lignocellulosic biomass by 40% following alkaline pretreatment without affecting plant growth as compared to wild type plants[73].
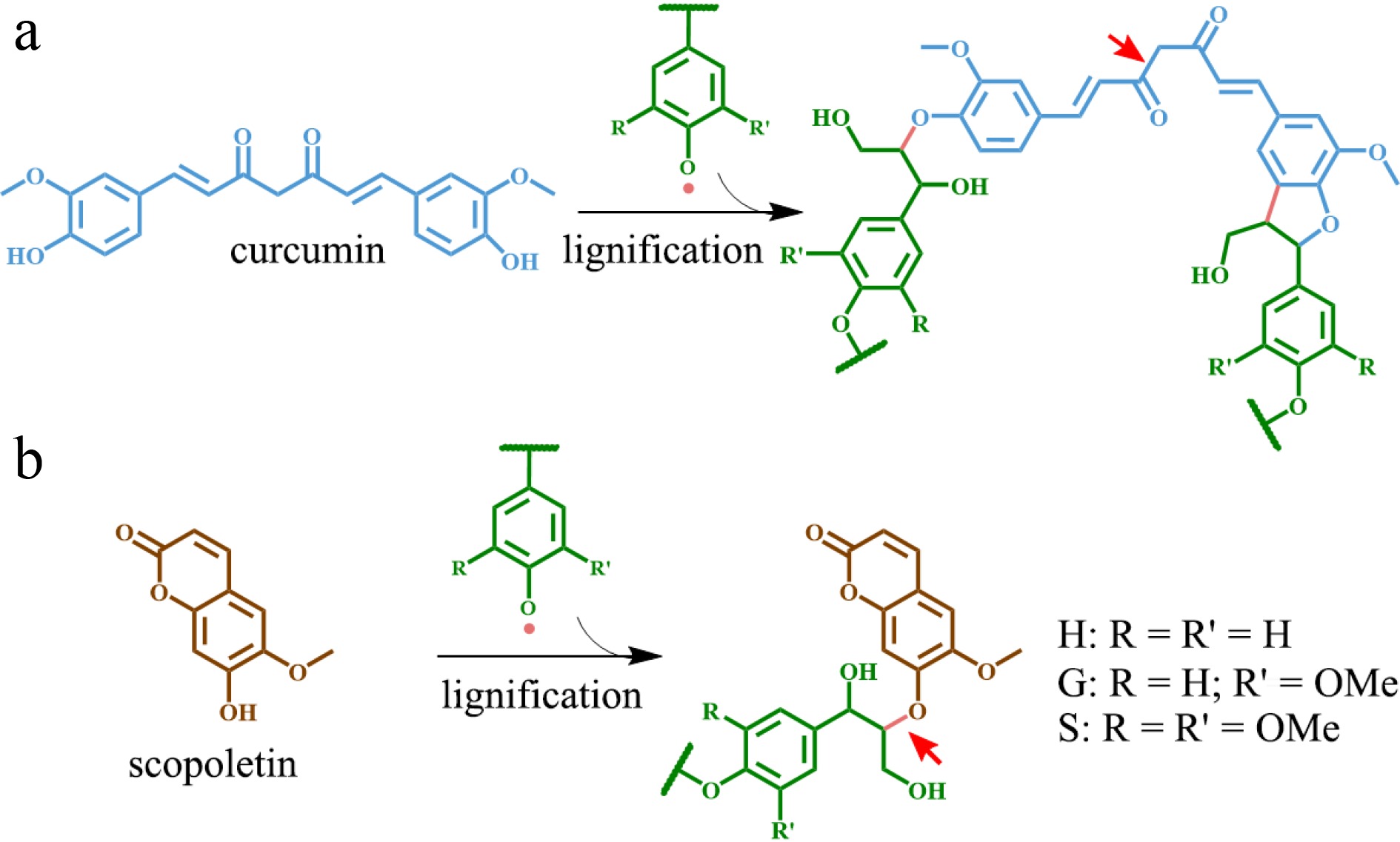
Figure 2.
Phenolic metabolites functioning as alternative monomers in lignin polymer. (a) Curcumin and the resulting lignin structure. (b) Scopoletin and corresponding lignin structure. The red arrow indicates the alkali-labile linkage on lignin.
Understanding alternative monomers biosynthesis and metabolism in plants as well as careful design are prerequisites for the successful engineering of non-native lignin. Following the successful integration into Arabidopsis lignin polymers, curcumin was also produced and incorporated into poplar lignin via heterologous expression of both DCS and CURS2 driven by the secondary cell wall CELLULOSE SYNTHASE A8-B promoter (ProCesA8-B). The transgenic trees exhibited shoot-tip necrosis and yield penalties without improving the saccharification yield[74], demonstrating that phenolic metabolism differs among species and that the strategy and knowledge acquired in model plants must be re-evaluated before being applied to crops. Additionally, some non-native lignin may be discovered to exist as authentic lignin in some tissues of certain plants in the future. Therefore, deep insights into unconventional lignin biosynthesis and regulation in planta may guide us to rationally design and engineer alternative lignin in different crops.
-
Plant lignin structural engineering research is still in its infancy and often encounters multiple challenges. Perturbances in the structure and content of lignin often inhibit plant growth and development. Despite experimental success stories, there can be possible unexpected results for the production of engineered lignin in field-grown trees on a commercial scale. Additionally, chemical modifications in lignin structure can exert toxic effects on plant health and the ecosystem. Some other challenges associated with engineered lignin may include the limited biodegradability in soil by microorganisms, the unpredictable lifecycle of resulting bioproducts, and environment-related regulatory standards. Therefore, careful designing of lignin monomers and a precise fine-tuning of lignin structure are necessary to minimize its adverse effects on plant growth as well as the environment.
The plant biologists pursued novel approaches for tailoring lignin to improve lignocellulosic biomass utilization without drastically altering lignin functioning in plants. The type and quantity of lignin monomers define lignin structure and are critical factors for redesigning lignin. Despite the remarkable flexibility of the lignification process for diverse phenolics in vitro, only some of them were suitable for lignin modification in planta[71]. Probably, we can let the plant's innate lignification mechanism choose the most suitable monomers for lignin engineering. Intriguingly, a vast number of phenolic compounds have been shown to behave as unconventional monomers that can integrate into lignin via radical coupling reactions. These native unconventional lignin monomers reshape the structure of lignin by offering a variety of functional groups and inter-unit linkages to lignin polymer. In the future, targeting these unconventional monomers may provide novel strategies to minimize the impacts of engineered lignin on plant development while improving the utilization efficiency of lignocellulosic biomass.
-
The authors confirm their contribution to the paper as follows: study conception and design: Zhao Y; data collection: Zhao Y, Abid M, Xie X, Fu Y, Huang Y, Cai Z, Lin H; draft manuscript preparation: Zhao Y, Abid M, Xie X, Fu Y, Huang Y, Cai Z, Lin H. All authors reviewed the results and approved the final version of the manuscript.
-
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
This work was supported by grants from the National Nature Science Foundation of China (Grant No. 32250011), The Chinese Academy of Sciences Project for Young Scientists in Basic Research (Grant No. YSBR-089), Distinguished Young Scholars of the National Natural Science Foundation of China (Overseas) and Shanghai Pujiang Program (Grant No. 22PJ1414300).
-
The authors declare that they have no conflict of interest.
- Copyright: © 2024 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Zhao Y, Abid M, Xie X, Fu Y, Huang Y, et al. 2024. Harnessing unconventional monomers to tailor lignin structures for lignocellulosic biomass valorization. Forestry Research 4: e004 doi: 10.48130/forres-0024-0001
Harnessing unconventional monomers to tailor lignin structures for lignocellulosic biomass valorization
- Received: 24 October 2023
- Revised: 27 December 2023
- Accepted: 12 January 2024
- Published online: 31 January 2024
Abstract: Lignin is an integral component in the plant secondary cell wall that imparts structural rigidity and integrity to plant tissues, contributes to the formation of the plant vascular system, and provides stress resilience. However, the heterogeneity and recalcitrance of lignin greatly impede the effective utilization of lignocellulosic biomass which represents the most abundant and valuable biogenic source for green and sustainable bioproducts. A customized lignin structure has the potential to alter lignin physicochemical properties and improve biomass utilization. Recently, researchers have reported that lignin polymerization can employ monomers beyond the traditional three monomers. The incorporation of these unconventional monomers can alter the physicochemical properties of lignin polymer by imparting lignin with a whole new structure. Here, we review the plant engineering efforts to alter lignin content, composition, and structure by harnessing non-conventional monomers. Rational manipulation of lignin structure with these unconventional lignin monomers may provide new strategies for minimizing the impacts of engineered lignin on plant growth and development meanwhile improving the utilization efficiency of lignocellulosic biomass.













