-
"Quzhiqiao" is immature fruit of Citrus paradisi 'Changshan Huyou', which is derived from Citrus × aurantium L.[1] and is widely cultivated in Changshan County (Zhejiang Province, China). As documented in the 'Standards for the Preparation of Traditional Chinese Medicine in Zhejiang Province' (2015 edition), the primary pharmacologically active compounds of "Quzhiqiao" are flavonoids, with naringin and neohesperidin serving as markers for assessing the quality of "Quzhiqiao"[2]. Furthermore, flavonoid compounds in "Quzhiqiao" also include components such as hesperidin, naringenin, hydrated hesperetin, hesperetin, luteolin, sinensetin and tangeretin, etc. However, extracting flavonoids from "Quzhiqiao" was challenging due to its hydrophobic character[3]. Due to the abundant flavonoid content, significant antioxidant activity, and valuable applications of flavonoids in "Quzhiqiao"[1,4], this study focuses on exploring the extraction of flavonoids using innovative extractant like octenylsuccinic acid (OSA) modified starch.
Until now, while there have been various methods for flavonoid extraction, the majority have primarily focused on solvent extraction[5−6]. However, this method tends to be time-consuming, and the solvents used can be costly and environmentally unfriendly[7]. Due to these factors, in recent years researchers have shifted toward using more efficient, environmentally friendly, and food-safe extraction materials. Microwave- and ultrasonic-assisted methods are known for providing thermal energy rapidly[8]. These methods can quickly disrupt the cell wall structure of plant cells, thereby enhancing the mobility and solubility of flavonoid molecules in water-based solutions[9−10].
OSA-modified starch, an organic compound based on starch, is considered safe for consumption[11]. It can be prepared by the esterification reaction between octenyl succinate glucoside and polyhydroxyl groups present on the starch surface[12]. Starch is a polymeric material. The gelatinization of OSA-modified starch can lead to an undesirable viscosity in the extract, which potentially affects the release rate of flavonoids from plant cells. To address this, altering the molecular weight of the OSA-modified starch has been proposed to adjust the viscosity and make it more suitable for flavonoid extraction[13]. Notably, the molecular weight of starch molecules can significantly affect their structure, modify their emulsification properties, and influence their extraction efficiency[14]. Hence, OSA-modified starch is regarded as a potential polymer material for extraction.
In this research, a novel extraction strategy was explored by employing OSA-modified starch with different molecular weights to extract flavonoids from the immature fruit of C. paradisi 'Changshan Huyou', also known as "Quzhiqiao". Through meticulous optimization, the study determined the ideal conditions for the extraction of flavonoids, including the concentration of the starch, ultrasonic power and duration, and the ratio of the mass of "Quzhiqiao" to the volume of gelatinized OSA-modified starch. Significant variations in antioxidant activities were observed across the extract solutions derived from OSA-modified starches of different molecular weights, suggesting possible compositional differences. This groundbreaking approach not only offers a refined method for flavonoid extraction but also provides valuable insights into the antioxidant properties of the extracts based on starch molecular weight. Meanwhile, this study would provide a data base for the promotion of OSA-modified starch in extraction techniques and the application of its extract solution in functional beverages.
-
"Quzhiqiao" was purchased from Changshan County Hongchun Fruit Professional Cooperative (Changshan, China). Waxy corn starch with amylopectin content ≥ 97.5% produced by Kunlun Biochemical Co., Ltd. (Gansu, China) and plant flavonoids assay kit produced by Beijing Solarbio Science & Technology Co., Ltd. (Beijing, China) were used. Octenyl succinic anhydride with purity ≥ 99.5% was from Jinan Haohua Industrial Co., Ltd. (Shandong, China) while β-amylase (7 × 105 U/mL in enzyme activity) used was from Yuanye Biotechnology Co., Ltd. (Shanghai, China).
Preparation of OSA-modified starches with different molecular weights
-
To prepare OSA-modified starch with different molecular weights, starch with different molecular weights was first obtained by enzymatic digestion (addition of 0%, 0.5%, 1%, 1.5%, and 2% β-amylase), and then esterified by adding OSA at a concentration of 3%. Xiang et al. described a method for preparing starches with different molecular weights and their esterified substances[15]. The prepared OSA-modified starches with different molecular weights were named Hst-OSAS, H-OSAS, M-OSAS, L-OSAS, and Lst-OSAS in the order of highest to lowest molecular weight.
Optimization of the extraction process
Sample preparation of "Quzhiqiao"
-
"Quzhiqiao" raw material was torn into small pieces to facilitate the release of flavonoids. They were washed with distilled water, laid on a plate and kept in an oven (Jinghong, DNP-9162, China) at 45 °C for 72 h to dry they were shriveled and crisp as moisture lost. The dried "Quzhiqiao" pieces were then ground into powder using a high-speed blender (Midea, MJ-PB80Easy218, China) and sieved using 40 mesh. The fine powder which passed through the mesh was referred to as "Quzhiqiao" powder. This powder was kept in an airtight desiccator before use.
Optimization of extraction factors
-
The primary factors subject to optimization in this study were the extraction methods, including stirring, ultrasonic-assisted extraction, and microwave-assisted extraction. The extraction mixtures were prepared under consistent conditions before using these extraction methods. An equal mass of OSA-modified starch with varying molecular weights was dispersed in deionized water (20 mL) and heated in a water bath at 75 °C with stirring (350 r/min) for 10 min until complete gelatinization was achieved. After cooling these solutions to room temperature (25 °C), an equal amount of "Quzhiqiao" powder was added and thoroughly mixed. The mixtures were stored in a dark place for 24 h to ensure that the "Quzhiqiao" powder was well distributed and interacted with the OSA-modified starch gelatinized solution. After 24 h, the mixtures were stirred again and followed by heating in a microwave. The extraction solutions were obtained after centrifugation (7,000 r/min) for 10 min.
Additionally, the single-factor method was applied to refine the extraction process and enhance the yield. Factors included extraction conditions (specifically extraction time), the solid-to-liquid ratio ("Quzhiqiao" powder to deionized water), the molecular weight of the OSA-modified starch, and the concentration of its solution. Details regarding the optimization conditions can be found in Table 1. All sample optimizations utilized yield as the primary indicator, with the yield testing methodology detailed in the following section.
Table 1. Optimization factors and indicators of the extraction process.
Extraction methods* Factors in the extraction process** Stirring Ultrasonic-assisted extraction Microwave-assisted extraction Extraction time (min) Ratio of solid to liquid (mg/mL) Concentration of OSA-modified starch (mg/mL) Indicators and their values Stirring speed:
500 r/min;
Extraction time:
40 minUltrasonic intensity:
1000 W;
Extraction time:
40 minMicrowave intensity:
1,000 W;
Extraction time:
5 min10 10 5 20 15 10 30 20 15 40 25 20 50 30 25 * The experiments were designed by the single-factor method and replicated three times; the extraction was carried out with the medium molecular weight OSA-modified starch solution (15 mg/mL) and a solid-liquid ratio of 20 mg/mL. ** The single-factor experiments were used under optimal extraction conditions. Medium molecular weight of OSA-modified starch was used. One of the variables was optimized, and the others were constant. Evaluation of extraction results
-
The extraction performance in this study was assessed based on yield, aiming to determine the optimal extraction method, the conditions, and the most appropriate molecular weight for OSAS extraction. The yield was calculated according to Eqn (1).
${\rm {Yield}}\; ({\text%})=\dfrac{C_{S} \times V_{S}}{m_{0}} \times 100 $ (1) m0 represents the initial mass of "Quzhiqiao" powder in each experiment, while Cs and Vs denote the concentration and total volume of the flavonoid extract, respectively, after centrifugation (25 mL).
To calculate the yield, the Cs from each experiment needs to be detected respectively, and the methods for the detection of total flavonoids in the prepared samples are shown below.
For precise determination of Cs, the extract solutions were concentrated using a rotary evaporator, yielding the extraction infusions. These infusions underwent ultrasonic extraction (1,000 W, 30 min) with anhydrous ethanol. The previous procedures mentioned were repeated in triplicate. Following centrifugation (5,000 r/min, 10 min), the total flavonoid was isolated from the infusions and any residual OSA was removed. The supernatants were collected and quantified using a 20 mL volumetric flask filled with anhydrous ethanol. All samples were subsequently assayed for their total flavonoid content, referencing naringin and employing the plant flavonoid assay kit.
Physiochemical properties of OSA-modified starches with different molecular weights
Molecular weight
-
The method for determining molecular weights was carried out as reported by Xiang et al.[15]. The molecular weight of starch was gauged using size exclusion chromatography on an Agilent 1100 HPLC system, equipped with a G1362A differential refractive index detector (RID) and a TSK G3000 PWxl gel column (300 mm × 7.8 mm, 7 μm). The reference standards are glucans with molecular weights ranging from 1 to 20 × 104 Da.
Degree of substitution
-
The emulsifying property of OSA-modified starch was determined by its degree of substitution. The method for determining the DS of OSA-modified starch is reported by Lopez-Silva et al.[16]. The OSA-modified starch (1 g) was dispersed in a 2.5 N HCl/isopropyl alcohol solution (20 mL) and stirred (350 r/min) for 30 min at room temperature (25 °C). 12.5 mL of 0.1 M hydrochloric acid solution was then added to this mixture, stirring at 350 r/min for 30 min. Subsequently, the sample was centrifuged (Thermo Scientific Heraeus Megafuge 11R) at 350 r/min for 10 min. The precipitate was washed with ethanol (90%) twice (5 mL × 2) and subsequently with distilled water six times (5 mL × 6) until no Cl− could be detected (using 0.1 M AgNO3 solution). The starch was resuspended in 150 mL of distilled water, placed in a boiling water bath at 350 r/min for 10 min, and then cooled to room temperature. The DS can be calculated using Eqn (2):
$ D S=\frac{\left(0.162 \times \dfrac{A \times M}{W}\right)}{1-\left(0.210 \times \dfrac{A \times M}{W}\right)} \times 100 $ (2) where A is the titration volume of the NaOH solution (mL), M is the molarity of the NaOH solution, and W is the OSA-modified starch's dry weight (g). The value 0.162 is the molecular weight of the glucosyl unit, and 0.210 are the molecular weight of the octenyl succinate group, unit in mol/g.
Viscosity detection of OSA-modified starches
-
Two grams of enzymatically dehydrolyzed OSA-modified starch were dispersed in 100 mL of distilled water, heated in a 75 °C water bath (JTLIANGYOU, SHJ-2AB, China) for 30 min, and continuously stirred using a magnetic stirrer. They were then cooled to room temperature and followed by viscosity analysis with a viscometer (Techcomp SNB-4, China) with a #2 spindle at 12 r/min at a constant temperature of 26 °C.
Fourier transform infrared spectroscopy detection
-
For FT-IR sample preparation, 1 mg of each of enzymatically dehydrolyzed OSA-modified starch sample (with enzyme additions of 0%, 0.5%, 1%, 1.5%, and 2%) was ground separately with 150 mg of KBr. After equilibrating for 24 h in an oven at 40 °C, the mixture was compressed. The range of scanning wavelength was from 400 to 4,000 cm−1 at a resolution of 4 cm−1[6].
In vitro antioxidant capacity assessment
Scavenging activity against the DPPH radical
-
The DPPH (2,2-diphenyl-1-picryl-hydrazyl-hydrate) free radical method is an antioxidant assay. It is based on the properties of the odd electron of the DPPH radical, which has a strong absorption at 517 nm, producing a violet color in ethanol. This free radical can be reduced in the presence of antioxidants and cause color changes. A higher DPPH radical scavenging activity indicates a higher antioxidant activity.
DPPH-ethanol solution (80 μM) was prepared. A total of 1.5 mL sample extract solution and 1.5 mL DPPH-ethanol solution were mixed and kept in a dark place for 30 min at room temperature (25 °C). The sample solution was assayed at 517 nm and ethanol was used as a control. Calculation refers to Eqn (3):
$ D P P H \; { radical \;scavenging\; activity }\;({\text%})=\left(1-\dfrac{A_{1}}{A_{0}}\right) \times 100 $ (3) where A0 is the absorbance value of the blank (1.5 mL of ethanol plus 1.5 mL of DPPH-ethanol solution); A1 is the absorbance value of the sample extract solution.
Scavenging activity against the ABTS radical
-
Trolox, a water-soluble analogue of vitamin E, is used as an antioxidant control standard. In ABTS assay, the oxidation of 2,2'-azinobis (3-ethylbenzothiazoline-6-sulfonic acid; ABTS) generates the colored free radical cation ABTS+. This radical rapidly reacts with antioxidants, leading to its reduction and discoloration. A standard Trolox solution was used in calculating the extent of decolorization[17].
Ferric reducing antioxidant power (FRAP) analysis
-
The principle of FRAP is that antioxidants can reduce the ferric 2,4,6-tripyridyl-s-triazine complex (Fe3+-TPTZ) to produce the blue Fe2+-TPTZ under acidic conditions (pH 3.6). The absorbance of the blue Fe2+-TPTZ at 593 nm is measured to determine the sample's total antioxidant capacity. The antioxidant capacity in this experiment was expressed in terms of the concentration of FeSO4 standard solution[18].
The reducing power determination was modified from the method reported by Nath et al.[19]. The working FRAP reagent was prepared by mixing 10 mL acetate buffer (0.3 mol/L), 1 mL of 0.01 mol/L TPTZ (2,4,6-tripyridyl-s-triazine) in 0.04 mol/L hydrochloric acid and 1 mL Ferric chloride (0.02 mol/L). A 0.1 mL sample was added to 3.9 mL of the freshly prepared FRAP reagent, thoroughly mixed. The absorbance was measured at 593 nm using an UV-VIS Spectrophotometer. A standard curve was prepared using different concentrations of FeSO4 solution. The reducing power of ferric was marked by the concentration of FeSO4 standard solution.
Data analysis
-
Certain experiments were repeated multiple times. The results were analyzed by GraphPad Prism (Version 9.3.1) and Excel 2010. All statistical data in this study were analyzed using SPSS 17.0 and Excel 2019.
-
OSA-modified starch with different molecular weights was prepared by adding different amounts of β-amylase. β-amylase functions as an exoenzyme, whereas α-amylase acts as an endoenzyme. It catalyzes the hydrolysis of α-1,4 linkage of polysaccharides into small molecules from its non-reducing end, while α-amylase can act at random sites of polysaccharides[20]. The reaction rate of β-amylase is slower than that of α-amylase. As shown in Table 2, the molecular weight of OSA-modified starch with different molecular weights was 20 × 104 Da for the highest molecular weight OSA-modified starch (Hst-OSAS), 12.51 × 104 Da for high molecular weight OSA-modified starch (H-OSAS), 9.25 × 104 Da for medium molecular weight OSA-modified starch (M-OSAS), 2.34 × 104 Da for low molecular weight OSA-modified starch (L-OSAS), and 1.61 × 104 Da for lowest molecular weight OSA-modified starch (Lst-OSAS), respectively. As more β-amylase is added, the molecular weight of the OSA-modified starch decreases.
Table 2. Molecular weights, degree of substitution, and viscosity of OSA-modified starches.
Sample Molecular weight (104 Da) DS (×103) Viscosity (Pa·s) Hst-OSAS 20.00 2.49 1.196 H-OSAS 12.51 2.51 0.921 M-OSAS 9.25 2.53 0.528 L-OSAS 2.34 2.57 0.444 Lst-OSAS 1.61 2.60 0.386 Notation of the molecular weight was as follows: Hst-OSAS, highest molecular weight OSA-modified starch; H-OSAS, high molecular weight OSA-modified starch; M-OSAS, medium molecular weight OSA-modified starch; L-OSAS, low molecular weight OSA-modified starch; Lst-OSAS, lowest molecular weight OSA-modified starch. The degree of substitution (DS) of different molecular weights of OSA-modified starch has been determined. The degree of substitution of these five different molecular weights of OSA-modified starch (Hst-OSAS, H-OSAS, M-OSAS, L-OSAS, Lst-OSAS) was 0.00249, 0.00251, 0.00253, 0.00257, 0.00260, respectively (Table 2). A lower molecular weight of OSA-modified starch correlates with a higher DS value. The probable reason for obtaining such results is that the low molecular weight of starch has a relatively large specific surface area. Therefore, the lower molecular weight of starch has a greater surface area for the substitution of OSA groups[21]. Hence, it would be essential to repeat the experiments to draw conclusive results and confirm that the DS increases as the molecular weight of OSA-modified starch decreases.
Viscosity result of OSA-modified starches
-
Viscosity analyses of OSA-modified starch revealed a correlation between the molecular weight of starch and its viscosity. Previous reports suggest that OSA-modified starches with higher viscosities are more resistant to mixing[22]. Consequently, this affects the flavonoid incorporation into gelatinised OSA-modified starch. Viscosity measurements were conducted at a consistent temperature of 26 °C. As depicted in Table 2, a higher molecular weight in OSA-modified starch corresponds to increased viscosity. Earlier research indicated that reduced β-amylase addition leads to a rise in the molecular weight of OSA-modified starch[13]. This higher molecular weight causes the starch molecules to become more entangled during gelatinisation, resulting in an increased viscosity.
FT-IR
-
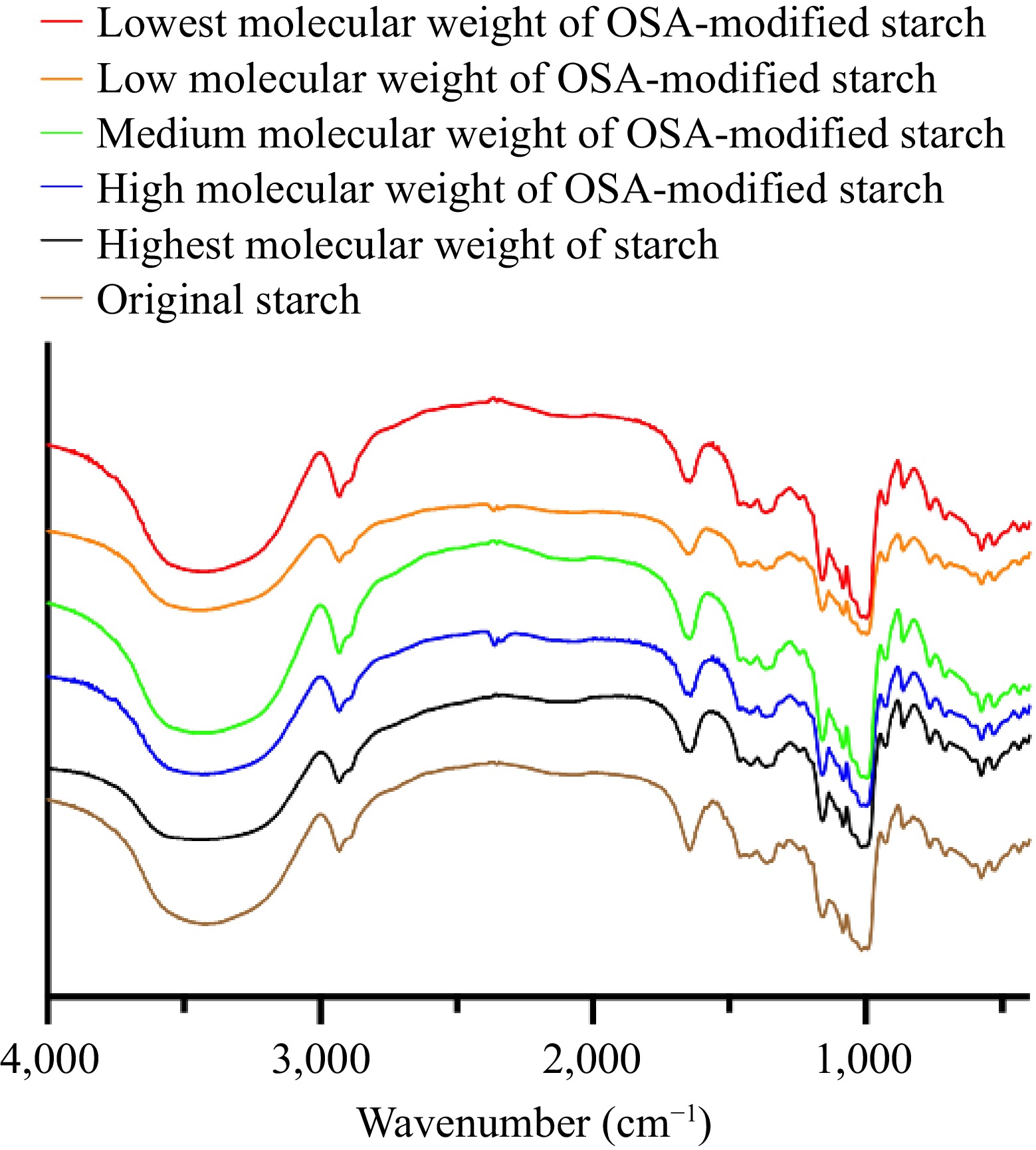
The FT-IR spectrum (Fig. 1) showed characteristic peaks at 1,724 cm−1 and 1,570 cm−1, confirming the success of the esterification reaction[23,24]. These peaks signify the introduction of the OSA group into the starch and the successful esterification of the starch particles. However, these peaks were not particularly prominent in this study's FT-IR spectra. The degree of substitution correlates with the intensity of these absorption peaks[25]. The subdued absorption peaks may be explained by low DS values in this study. Furthermore, variations in the molecular weights of OSA-modified starches had minimal impact since the DS values were closely aligned.
Optimization results of the extraction process
-
The single-factor method was employed to investigate potential factors influencing the yield during the extraction process, with results depicted in Fig. 2. During optimization, the extraction method and solvent type were refined based on emulsifying attributes and polarity alignment with the flavonoid molecule[26,27]. In combination with OSA-modified starch solution, ultrasound-assisted extraction methods provided superior results, establishing them as the optimal extraction method. The concentration of extraction solution and molecular weight of OSA-modified starch were considered in subsequent optimization, shown in Fig. 2c. As initial concentrations shifted, extraction efficiency varied with molecular weight, generally first declining and then increasing. Notably, medium molecular weight OSA-modified starch demonstrated peak performance at an initial concentration of 20 mg/mL . Further optimization, using the single-factor method, pinpointed the ideal solid-to-liquid ratio at 15 mg/mL and extraction duration at 40 min.
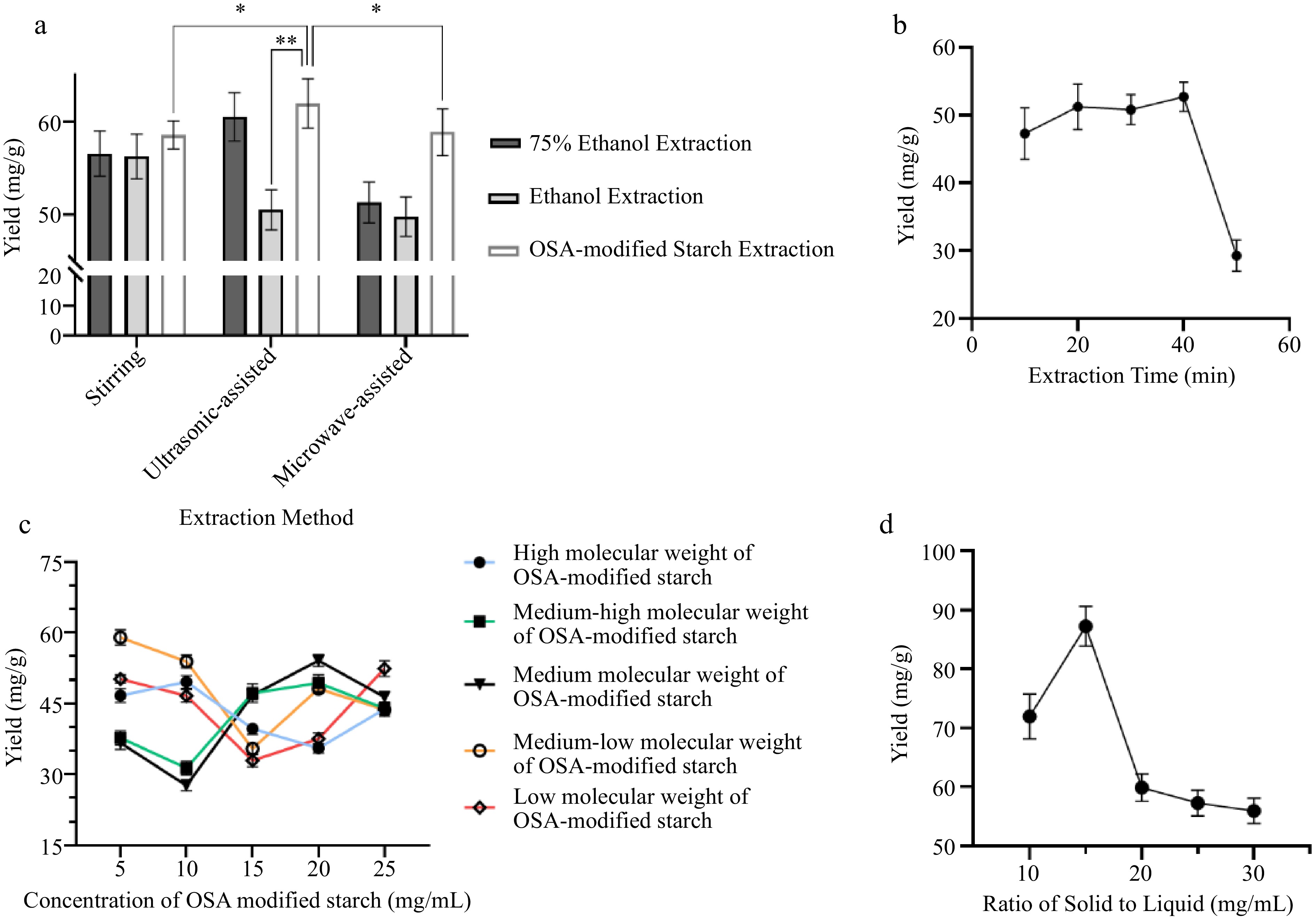
Figure 2.
Optimization results of extraction factors (extraction time, extraction method, ratio of solid to liquid, different molecular weights of OSA-modified starches).
The notable difference in flavonoid extraction rates using OSA-modified starch may stem from the viscosity and degree of esterification of the OSA-modified starch, as well as the self-assembly state of OSA-modified starch molecules in water induced by these properties. Current research findings suggest that a lower viscosity does not unequivocally result in a higher extraction rate, even though a reduced molecular weight might yield a higher degree of esterification. This indicates that both viscosity and degree of esterification are crucial determinants of the extraction rate, but their effects are not linear. Previous studies have shown that OSA-modified starch can self-assemble in water, forming internal cavities of varying dimensions and can establish supramolecular complexes with naringin. Concurrently, mid-molecular weight OSA-modified starch has been demonstrated to enhance the solubility of naringin in water more proficiently. Furthermore, in this study, naringin was utilized as a reference standard to calibrate the total flavonoid content[15]. Considering these aspects collectively, the optimal size of the supramolecular internal cavity of the OSA-modified starch during extraction, along with the solubilizing effect of OSA-modified starch on naringin, might be the primary factors driving the observed variations in extraction rates.
In summary, the optimal extraction process encompasses the following steps:
Firstly, disperse 800 mg of medium molecular weight OSA-modified starch (M-OSAS) in 40 mL of deionized water. After 24 h of sealed storage, heat and stir the mixture in a water bath at 350 r/min and 75 °C for 10 min. Subsequently, cool the gelatinized M-OSAS to room temperature.
Secondly, introduce 600 mg of 'Quzhiqiao' powder to the 40 mL dextrinized M-OSAS solution. After thorough mixing and sealing with cling film, subject it to ultrasonication (1,000 W, 40 min). Post-ultrasonication centrifugation produces the extraction solution.
thirdly, repeat the extraction solution process three times as per the yield test method detailed in Subsection "Evaluation of extraction results". The resultant yield from this optimized process is 63.72% ± 5.12%.
In vitro antioxidant capacity result of the extraction solution
-
Building on the optimal extraction process detailed in Subsection "Optimization results of the extraction process", flavonoids from "Quzhiqiao" powder were extracted using OSA-modified starches of varying molecular weights. The resultant extract solutions underwent in vitro antioxidant analysis, with findings summarized in Table 3. The DPPH radical scavenging activity of these extracts correlated positively with the molecular weight of OSA-modified starch, suggesting an increase in components soluble in organic systems as the molecular weight rises[28−29]. Notably, extracts from both M- and L-OSAS showcased strong ABTS radical scavenging activity, indicating a richer presence of water- and alcohol-soluble components[30−31]. The M-OSAS extract, in particular, displayed superior Fe3+ reduction capacity. Significant variations between results emphasize the substantial impact of OSAS molecular weight on Fe3+ reduction within its extract. These findings underscore the influence of OSAS molecular weight on its extraction composition, suggesting it could be a pivotal factor in its efficacy in extracting total flavonoids from "Quzhiqiao".
Table 3. In vitro antioxidant results of extraction solution.*
Sample DPPH radical scavenging activity (%) Equivalent to Trolox (μM)** Equivalent to FeSO4
(mM)Hst-OSAS 11.41 ± 0.77a 885.72 ± 3.21a 1.460 ± 0.048a H-OSAS 23.70 ± 3.43b 888.94 ± 8.32a 1.515 ± 0.026b M-OSAS 33.63 ± 0.45c 896.20 ± 9.86b 1.615 ± 0.067c L-OSAS 54.84 ± 4.94d 895.06 ± 8.63b 1.497 ± 0.034d Lst-OSAS 55.47 ± 3.85d 889.43 ± 7.92a 1.382 ± 0.018e * Each experiment was repeated three times and expressed as mean ± SD. Different letters in the same column represent a significant difference between the two groups of data (p < 0.05). ** Trolox, a water-soluble analog of vitamin E, is used as a control antioxidant standard. -
This study examined the extraction capabilities of OSA-modified starch, specifically targeting the flavonoid component in "Quzhiqiao". Optimal conditions were defined, revealing that a gelatinized solution of OSA-modified starch with a molecular weight of 9.25 × 104 Da was the most effective for flavonoid extraction from "Quzhiqiao" under ultrasound assistance (1,000 W, 40 min). It was also determined that a starch solution concentration of 20 mg/mL and a solid-to-liquid ratio of 15 mg/mL were optimal. Following these parameters, the flavonoid yield from "Quzhiqiao" using OSA-modified starch reached 63.72% ± 5.12%. Furthermore, OSA-modified starch extracts demonstrated significantly different antioxidant properties in vitro. These differences greatly affect the experimental data in different models of antioxidant experiments in vitro. As a result, molecular weight might play a crucial role in determining extraction efficiency and extracted ingredients.
-
The authors confirm contribution to the paper as follows: conceptualization, data curation, methodology, formal analysis, software, visualization, roles/Writing - original draft: Wang L, Zhou X; writing - review & editing: Wang L, Zhou X, Lu S, Cui Q; funding acquisition, project administration: Lu S, Cui Q; supervision: Liu J, Quek SY; resources, validation: Lu S, Cui Q, Liu J, Quek SY. All authors reviewed the results and approved the final version of the manuscript.
-
Due to administrative requirements, the original data of the experiments during the research period of the project are not available to the public, but available from the corresponding author or the first author upon request.
This work was supported by the National Natural Science Foundation of China (grant number 31571892), National Natural Science Fund of China (grant number 82204552), Natural Science Foundation of Zhejiang Province (grant number LQ22H280007), Research Project of Zhejiang Chinese Medical University (grant number 2022JKZKTS10).
-
The authors declare that they have no conflict of interest.
-
# These authors contributed equally: Lu Wang, Xuyi Zhou
- Copyright: © 2023 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Wang L, Zhou X, Lu S, Quek SY, Liu J, et al. 2023. Utilizing OSA-modified starch with various molecular weights for flavonoid extraction from "Quzhiqiao" (immature fruit of Citrus paradisi 'Changshan Huyou'). Beverage Plant Research 3:30 doi: 10.48130/BPR-2023-0030
Utilizing OSA-modified starch with various molecular weights for flavonoid extraction from "Quzhiqiao" (immature fruit of Citrus paradisi 'Changshan Huyou')
- Received: 30 June 2023
- Revised: 28 September 2023
- Accepted: 07 October 2023
- Published online: 10 November 2023
Abstract: In order to investigate the extraction efficiency of flavonoids by octenyl succinic acid-modified starch (OSA-modified starch) with varying molecular weights, the immature fruit of Citrus paradisi 'Changshan Huyou', commonly known as "Quzhiqiao", was used as the extraction target in this research. OSA-modified starch was successfully identified and refined, ensuring its molecular weight was appropriate and extraction was optimized. The experimental findings suggested that a concentration of 20 mg/mL of medium molecular weight gelatinized OSA-modified starch was optimal for extracting flavonoids from "Quzhiqiao" using an ultrasound-assisted method (1,000 W, 40 min). Additionally, the ratio of the mass of "Quzhiqiao" to the volume of gelatinized OSA-modified starch employed in this process was determined to be 15 mg/mL. The findings from the in vitro antioxidant studies indicated significant variations in the antioxidant activities of the extract solutions of OSA-modified starches with varying molecular weights. These differences can potentially be attributed to substantial variations in the extract solutions composition.
-
Key words:
- OSA-modified starch /
- Quzhiqiao /
- Extraction method /
- Flavonoids /
- Antioxidant