-
Protein homeostasis (proteostasis) in the endoplasmic reticulum (ER) is important not only for plant growth and development, but also for adaptations to various environmental challenges[1]. When the protein folding demands exceed the folding capability in ER, the proteostasis status becomes disrupted. To overcome this imbalance, the unfolded protein response (UPR) is triggered to decrease the accumulation of unfolded and misfolded proteins in the ER through activating downstream target genes[2]. Two canonical UPR pathways are identified in plants[3,4], and their roles in responses to environmental stresses as well as in growth and development have been extensivly studied over the past 30 years[5]. However, the role of UPR pathways in grain development in cereals has just begun to emerge.
The endosperms of cereal crops such as rice (Oryza sativa L.) account for most of the grain weight and are major resources for human food and animal feed[6]. Endosperm development is a complex process, involving a series of processes such as cell division and differentiation, transport and accumulation of photosynthetic products, and enrichment of stored substances. Among the storage products in the rice endosperm, starch and seed storage proteins (SSPs) are the major nutritional components, both of which are important for nourishing the embryo during embryogenesis and seed germination[6].
SSPs are synthesized and folded mainly in the ER in the secretory pathway. As early as 1991, it was found that an increase of luminal binding protein (BiP) protein, specifically in the endosperm of maize floury-2 (fl2) mutant, leads to the constitutive activation of UPR and a starchy endosperm phenotype with increased lysine content and reduced amount of prolamin (zein) proteins[7]. Later it was found that the accumulation of non-processed 24-kD ɑ-zein protein in the fl2 mutant is responsible for the observed floury phenotype, and overexpression of a mutated ɑ-zein protein with non-cleavable signal peptide reproduces the fl2 phenotype in transgenic maize[8]. Mutant analysis demonstrated that accumulation of a mutated 19-kD ɑ-zein also interferes with the proper assembly of zeins into protein bodies and upregulates several UPR downstream genes in the endosperms of the maize fl4 mutant[9]. These results indicate that proper folding of storage proteins in the ER is critical for grain quality. In this review, we summarize the canonical UPR pathways in rice, and focus on the recent advances on the understanding the role of UPR in SSP accumulation in rice grains, and identify some unanswered issues related to UPR and rice grain quality.
-
ER is the key organelle for processing and folding nascent polypeptide chains, and forms complex spatial structures in the secretory pathway. Protein folding in the ER is error-prone, and a set of quality control steps have evolved to minimize misfolded protein accumulation in the ER[1]. Among them, UPR pathways are activated to increase protein folding capacity and decrease misfolded protein accumulation[10]. The principles of UPR pathways are well conserved among eukaryotic organisms from yeast to worms, plants, flies and humans, but the protein sequences of some UPR components are different[5]. In the model plant Arabidopsis, at least two UPR pathways are identified, the AtS2P-AtbZIP28 pathway and the AtIRE1-AtbZIP60 pathway, in both of which membrane-associated transcription factors are involved[4]. Generally, the basic domain leucine zipper (bZIP) protein AtbZIP28 resides in ER membranes in a dormant form, with its C-terminal domain binding to the molecular chaperones BiPs in ER lumen[11−13]. Upon accumulation of misfolded proteins in ER, AtbZIP28 is dissociated from AtBiPs and translocated from ER to Golgi, where it is subjected to proteolysis executed by Golgi-localized site-specific proteases, such as SITE-2 PROTEASE (AtS2P)[14−17]. Then the activated N-terminal cytosolic part of AtbZIP28 having the DNA-binding domain and transcriptional activation domain, enters the nucleus to upregulate UPR downstream genes[18]. In contrast, AtbZIP60 is activated by unconventional splicing depending on the ER membrane-associated INOSITOL-REQUIRING ENZYME 1A/1B (AtIRE1A/1B), of which the N-terminal domains are located in the ER lumen sensing the accumulation of misfolded proteins in the ER[19]. Under normal conditions, AtbZIP60 has a transmembrane domain that resides in the ER membranes[20]. Upon ER stress, 23 bases in the double stem-loops of AtbZIP60 mRNA are spliced out by the activated AtIRE1A/1B, resulting in a reading frame shift and production of a newly translated AtbZIP60 without any obvious transmembrane domain[21]. AtbZIP60 is then translocated to the nucleus to activate UPR downstream genes[20]. Both AtbZIP28 and AtbZIP60 are required for the induction of UPR genes and ER stress tolerance in Arabidopsis[22,23].
The UPR pathways are well conserved in rice plants[24] (Fig. 1a). Briefly, OsbZIP60/OsbZIP16 is the ortholog of AtbZIP28 in rice, which is activated presumably through proteolysis[25]. In contrast, OsbZIP50/OsbZIP74 is the rice ortholog of AtbZIP60, it is activated by OsIRE1-dependent mRNA unconventional splicing[26,27]. The expression of OsIRE1 is negatively regulated by SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE 6 (SPL6), and down-regulation of OsSPL6 results in hyperactivation of UPR and cell death in rice panicles[28]. Both OsbZIP50 and OsbZIP60 recognize the pUPRE-II cis-element (GATGACGCGTAC), while OsbZIP60 specifically binds to the pERSE-I cis-element (CCAAT-N10-CACG)[25]. OsbZIP50 is also activated under heat stress conditions and involved in a regulatory circuit together with rice NAC with transmembrane motif 1 (NTM1)-like 3 (OsNTL3)[27,29,30].
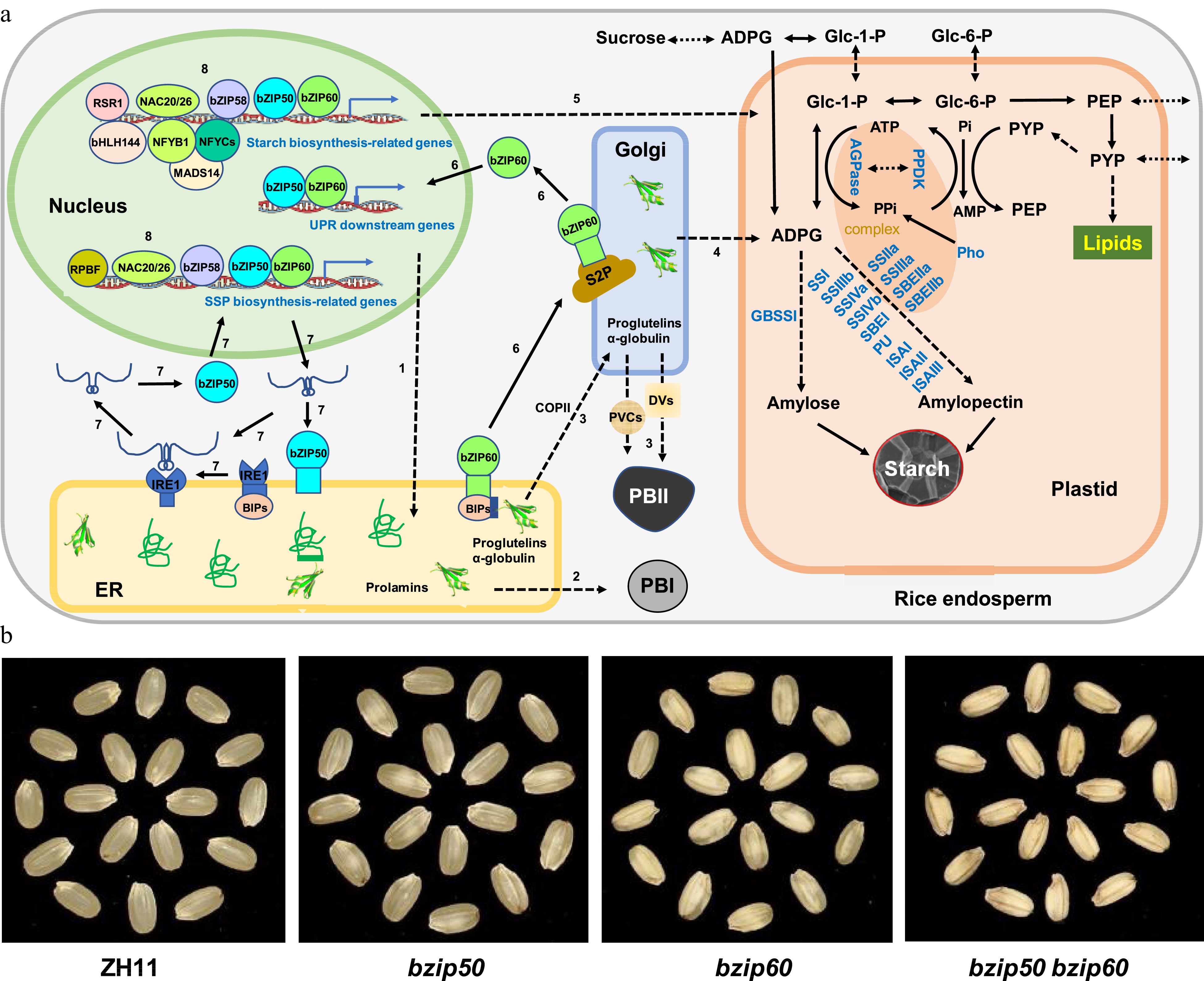
Figure 1.
Coordination of SSP and starch biosynthesis by the UPR pathways in rice endosperm. (a) Regulation of SSP and starch accumulation. Nascent polypeptides of SSPs with N-terminal signal peptide sequences are folded in the ER lumen (1), the properly folded prolamins are secreted and matured in PBI (2), while properly folded proglutelins and ɑ-globulin are exported from ER into Golgi via COPII vesicles and then to PBII mediated by DVs and PVCs (3). In contrast, proteins related to starch biosynthesis are transported either from Golgi to plastid (4) or from cytosol to plastid (5) to regulate starch biosynthesis. Two major UPR pathways, bZIP60-S2P and bZIP50-IRE1, are activated when ER protein homeostasis is disrupted in rice plants. Under such conditions, the ER membrane-associated transcription factor bZIP60 is disassociated with the chaperones BiPs, which results in the ER-to-Golgi transporting and processing of bZIP60 by Golgi-localized proteases such as S2P (6), the activated bZIP60 enters the nucleus and induces the expression of UPR downstream genes as well as SSP and starch biosynthesis-related genes (6). On the other hand, instead of encoding an ER membrane-associated transcription factor bZIP50, the bZIP50 mRNA is unconventionally spliced by the activated ER protein IRE1, leading to the open reading frame shift and a newly translated protein without any obvious transmembrane domain (7). The activated bZIP50 enters the nucleus to play a similar role as bZIP60 (7). Some other transcription factors, such as bZIP58 and NAC20/26, regulate the expression of genes involved in both SSP and starch biosynthesis (8). For protein abbreviations, see the text. (b) Phenotypic analysis. Wild-type (ZH11), single mutant of bZIP50 (bzip50) or bZIP60 (bzip60), and their double mutant (bzip50 bzip60) are grown under normal growth conditions and their dehulled seeds are photographed at maturity.
-
The endosperm is an important energy storage organ of cereals and one of the important food sources for human beings. Starch consists of more than 80% of the mature rice endosperm, and SSPs contain approximately 8%–10% of the dry weight of grains. There are two types of starch in rice endosperms, the linear amylose (AM) (10%–30%) and branched amylopectin (AP) (70%–90%)[6]. In contrast, rice SSPs are grouped into four categories based on their solubility: glutelin (acidic or basic solution-soluble), prolamine (alcohol-soluble), globulin (salt-soluble), and albumin (water-soluble)[31]. Both the total amount and the ratio between starch and SSPs are important for the seed quality of rice.
The metabolites of starch in endosperm is the key point for endosperm development and is a complex and coordinated process involving a series of key enzymes (Fig. 1a), which include adenosine diphosphate glucose pyrophosphorylase (AGPase), granule bound starch synthase I (GBSSI), soluble starch synthases (SSI, SSIIa, SSIIIa, SSIVa), as well as starch branching enzymes (SBEI, SBEIIa, SBEIIb), isoamylases (ISAI, ISAII and ISAIII), pullulanase (PU), and starch phosphorylase (Pho)[32]. Some of these enzymes interact with each other to form heteromeric protein complexes and are subject to post translational modifications[33]. The disruption of genes encoding these enzymes impairs endosperm development, resulting in the altered appearance of seeds or changed characteristics of starch. Some of those mutants showed floury endosperms while the others displayed chalky endosperms[34−37]. Recently, several transcript factors have been found to regulate starch synthesis in rice. For example, rice nuclear factor Y subunit B1 (OsNF-YB1) activates the expression of genes involved in starch accumulation either alone or together with other transcription factors such as RICE STARCH REGULATOR 1 (RSR1), OsNF-YC10, OsNF-YC12, rice basic helix-loop-helix 144 (OsbHLH144) and OsMADS14[38−42].
Glutelin and prolamine are two major SSPs in rice grains, and glutelin is more nutritious than other SSPs due to its relative high lysine content and digestibility. Rice SSPs are synthesized in the secretory pathway starting from the rough ER, and are then transported and deposited into the protein body I/II (PB-I/PB-II). The biosynthetic pathways of rice SSPs have been recently reviewed and will not be covered in the current paper[43]. Briefly, genes encoding rice SSPs are spatially and temporally expressed during endosperm development under the control of several transcription factors, leading to unique protein body formation and SSP deposition (Fig. 1a). The Opaque2 (O2)-like bZIP transcription factor RICE SEED bZIP1 (RISBZ1, also known as OsbZIP58), and the RICE P BOX BINDING FACTOR (RPBF) in the DNA-BINDING WITH ONE FINGER (DOF) transcription factor family are two major transcription factors for regulating SSP genes via recognizing the GCN4 motif [TGA(G/C)TCA] and P box (TGTAAAG), respectively[44,45]. Interestingly, RISBZ1(OsbZIP58) also controls starch biosynthesis by binding to the promoter of six genes involved in starch synthesis, i.e. AGP-like 3 (AGPL3), Waxy (Wx), SSIIa, SBE1, SBEIIb, and ISAII, to regulate their expression[45].
-
UPR could be induced during endosperm development due to the increased demand of SSP folding in the ER. The key chaperone gene involved in folding of secretory proteins, OsBiP1, is predominantly expressed during rice seed maturation, either suppression or over-expression of OsBiP1 not only resulted in altered intracellular structure of endosperm cells, but also affected SSP and starch accumulation[24,46]. OsERdj7 is an ER-resident J-domain-containing DnaJ protein and acts as a co-chaperone for Hsp70. Suppressing the expression of OsERdj7 in rice endosperms with RNAi technology reduces SPP accumulation[47]. Further, mutation of a rice protein disulphide isomerase-like 1-1 (OsPDIL1-1) gene involved in protein folding in the ER, triggers the expression of many UPR genes in the rice endosperm and produces small grains with floury endosperms[48]. These suggest that UPR is associated with endosperm development in rice grains, which is essential for SSP accumulation and rice endosperm development.
As mentioned above, OsbZIP60 and OsbZIP50 are two important UPR regulators in rice. Recently, Yang et al. reported that knock-out mutation of OsbZIP60 causes high grain chalkiness and produces aberrant structure with storage substances, while knock-out mutation of OsbZIP50 showed less chalkiness[49]. We generated the double loss-of-function mutant plant osbzip60 osbzip50 by crossing the respective single mutants[29], and the seeds of the double mutant are extremely shrunken and chalky (Fig. 1b), i.e. the phenotype of the double mutant is much more severe than the respective single mutants while the vegetative growth of these plants appears normal under standard growth conditions. The osbzip60 mutant grains have reduced amylose and protein content, and several genes related to grain chalkiness and protein synthesis such as GLUTELIN PRECURSOR OVERACCUMULATION 3 (GPA3), FLOURY SHRUNKEN ENDOSPERM 1 (FSE1), FLOURY ENDOSPERM 7 (FLO7), CHALKINESS 5 (Chalk5), OsNF-YB1 and rice PYRUVATE KINASE 2 (OsPK2), are down-regulated in the osbzip60 mutant[49]. These results strongly support that UPR is critical for starch and SSP accumulation in rice grains, which directly determines the quality of rice grains.
The UPR operates to minimize the accumulation of unfolded proteins in the ER by increasing the protein-folding and degradation capacity of the ER. Many genes involved in endoplasmic reticulum associated degradation (ERAD) are up-regulated during the UPR in plants[1]. The HMG-CoA REDUCTASE DEGRADATION 1 (HRD1) complex is one of the most important ERAD machineries in plants, and it contains OSTEOSARCOMA 9 (OS9) and HRD3, which recognizes misfolded ERAD clients and the E3 ligase HRD1 that polyubiquitinates the ERAD clients[50]. Endosperm-specific suppression of OsHrd3 reduces the level of polyubiquitinated proteins and enhanced UPR in rice endosperms, resulting in deformed PB-I formation with aberrant aggregates of prolamin[51]. Rice DERLIN-LIKE PROTEIN 1 (OsDER1), a conserved subunit of the HRD1 complex in plants, interacts with OsHRD1, OsHRD3, and the AAA-ATPase OsCDC48[52]. Suppression of OsDER1 results in floury, shrunken seeds in rice[52]. Therefore, ERAD is tightly associated with UPR to regulate grain development in rice plants.
-
Grain chalkiness affects the appearance, milling, cooking and eating quality of rice grains; therefore, it is an undesirable trait for rice production. Intriguingly, both overexpression and mutation of OsbZIP60 leads to chalky endosperms[49]. This raised the question as to what is the optimial UPR level for ideal endosperm development. Interestingly, there are natural variations in the promoter and coding regions of OsbZIP50 and OsbZIP60[49]. Thus, in the future it will be worthy to try to optimize UPR level for breeding high quality rice. In addition, a prime-editing-library-mediated saturation mutagenesis method is recently developed and successfully used to generate value-added rice plants[53]. Therefore, direct evolution of genes encoding key UPR regulators with this new technology are needed in the future for rice grain quality improvement.
Starch is stored in the starch granule, which has a complex structure with a hierarchical order composed of two distinct types of glucose polymer, i.e. amylose and amylopectin. In contrast, SSP biosynthesis starts from ER, passes through ER-Golgi-protein storage vacuoles (PSVs) routes, or directly buds off ER, and maturated in PB-I or PB-II. One unanswered question is how starch biosynthesis and SSP accumulation in rice grains are coordinated during endosperm development. Previous work has shown that OsNAC20 and OsNAC26 functions redundantly in starch and SSP accumulation through activating the expression of α-globulin, GluA1, GluB4/5, Pu, SSI, and 16 kD prolamin[54]. Recently, OsbZIP60 has been demonstrated to regulate genes related to grain chalkiness and protein synthesis, and OsbZIP50 is activated in the osbzip60 mutant plants[49]. Future studies on the starch and SSP biosynthesis in the osbzip60 osbzip50 double mutant plants is needed to further understand how the biosynthesis of starch and SPPs is balanced in rice endosperms.
In conclusion, lines of evidence support that proper folding of ER proteins are required for seed development and grain quality in rice. ER protein homeostasis safeguarded by the UPR pathways plays critical roles in the coordination of starch and SSP accumulation in rice endosperm.
We apologize to researchers whose work has not been cited in the review due to space constraints. This project was financially supported by grants from State Key Project of Research and Development Plan (2021YFF1000400), National Natural Science Foundation of China (32072038), and Zhejiang Provincial Talent Program (2019R52005).
-
The authors declare that they have no conflict of interest.
- Copyright: © 2022 by the author(s). Published by Maximum Academic Press on behalf of Hainan Yazhou Bay Seed Laboratory. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Zhou YF, Qing T, Shu XL, Liu JX. 2022. Unfolded protein response and storage product accumulation in rice grains. Seed Biology 1:4 doi: 10.48130/SeedBio-2022-0004
Unfolded protein response and storage product accumulation in rice grains
- Received: 30 May 2022
- Accepted: 19 August 2022
- Published online: 06 September 2022
Abstract: Secretory and transmembrane proteins start to synthesize and fold in the endoplasmic reticulum (ER). When the balance between protein folding demands and protein folding capability in the ER is broken, a well-conserved process known as the unfolded protein response (UPR) is induced to restore protein homeostasis. The grain quality of rice (Oryza sativa L.), one of the most important crops that feed more than half of the world’s population, is determined by the accumulation of nutritional components, such as seed storage proteins (SSPs) and starches in the grains. Rice SSPs are synthesized in the secretory pathways of endosperms and their biosynthesis is subject to complex regulation. Here, we focus on summarizing recent advances in our understanding of the role of UPR in grain development, especially in SSP biosynthesis in rice, and provide future perspectives on unanswered questions on improving grain quality through modulating UPR in rice.
-
Key words:
- grain quality /
- Oryza sativa /
- protein homeostasis /
- seed storage proteins /
- unfolded protein response













