-
Coffee is one of the viable agricultural commodities making an economic and social impact worldwide. Many coffee-producing countries in the coffee belt are benefiting from this crop because it represents a major source of income[1]. The global consumption of coffee is expected to rise from 800,000 - 60 kg bag to 167.9 million - 60 kg bag, by which the European Union, USA and Brazil have the largest gains[2]. The Philippines is only 15% self-sufficient in coffee despite having the ideal conditions to grow quality coffee varieties[3]. Recently, the demand for quality coffee has increased because of the available specialty coffees in the market[4]. The coffee quality is majorly associated with post-harvest management operations that determine the cupping quality profiles, this includes the wet processing method[5].
Fermentation is a crucial step in wet processing and it is commonly driven by microorganisms that can play a number of roles, such as degradation of mucilage from the coffee parchment, inhibition of mycotoxin-producing fungi, and production of flavor-active compounds[6]. The coffee mucilage has sticky pectin polysaccharide substances that are laborious to eliminate using water. Natural plant enzymes may facilitate the degradation of mucilage, however it is not a sufficient process. The participation of microorganisms like pectinolytic bacteria can lead to the complete degradation of pectinaceous substances, accelerating the production of green coffee beans[7]. The different metabolites and organic acids produced by pectinolytic bacteria are stored in the coffee beans and may affect the coffee quality[5]. Thus, the role of microbes to control fermentation and promote specialty coffee has been extensively studied by utilizing them as starter cultures[7].
Starter culture application aims to accelerate the fermentation of coffee mucilage using microbes like yeast and bacteria[6]. These starter cultures can rapidly increase the acidity by reducing the pH and shorten the fermentation time period. Bacterial starters are producers of acid compounds and yeast starters of volatile alcohols[4]. As a result, they can produce coffee with unique aromas and flavors, leading to new perspectives of coffee quality[8].
In this study, we aimed to characterize and identify indigenous pectinolytic bacteria isolated from wet coffee fermentation for their potential use as starter cultures. The hydrolytic enzyme production of these pectinolytic bacteria were evaluated along with sugar fermentation and tolerance to a wide range of abiotic stresses.
-
A total of 2 kg of cherries from Arabica trees (Typica) were collected from a coffee farm in Itogon, Benguet (1,253.701 masl; Lat. 16.34056465° N and Lon. 120.62705884° E). Ripe cherries were manually picked, washed, and immediately soaked in water to separate the defects (i.e., overripe, undeveloped, and infected coffee cherries). After 12 h, sinkers were manually depulped and the parchment coffee with mucilage was subjected to 48-h fermentation in a controlled environment. The pH and temperature of the fermentation setup were monitored every 12-h interval sampling period.
Enumeration and isolation of aerobic bacteria
-
During fermentation, the colony forming units (CFU) mL−1 of the total aerobic bacteria were enumerated, following the serial dilution and spread-plate method. A total of 10 mL liquid fraction from 12-h interval time points (i.e., 0, 12, 24, 36, and 48 h) during wet fermentation were collected and diluted in 90 mL sterile saline solution (0.85% NaCl [w/v]). The mixture was serially diluted 10-fold up to 10−5 dilution and immediately plated on the plate count agar (PCA) medium in triplicates. Agar plates were incubated at room temperature for 24 h and the CFU mL−1 was calculated.
A total of 35 bacterial colonies were randomly picked based on their unique colony morphologies and were subjected to purification through successive streaking on PCA medium. The purity of the cultures was confirmed through Gram staining and pure cultures were stored at 4 °C until further use.
Screening of pectinolytic bacteria
-
The bacterial isolates were screened for their ability to produce pectinase using a pectin agar medium, containing 0.5 g·L−1 peptone, 0.3 g·L−1 beef extract, 0.5 g·L−1 NaCl, 4 g·L−1 citrus pectin, and 12 g·L−1 agar. Initial number of cells were adjusted to 0.5 McFarland standard that is approximately 1.5 × 108 CFU mL−1, this was used in all experimental plate assays. Briefly, a total of 4 μl of 24-h broth culture inoculum was spot inoculated in triplicate on the citrus pectin agar medium and plates were incubated at 30 °C for 96 h. Pectinase activity was confirmed based on the appearance of clear zones around colonies after flooding of 300 mM potassium iodide-iodine solution. The pectin solubilization index was calculated by dividing the colony diameter + zone diameter over colony diameter.
Pectinase enzyme assay
-
Pectinolytic bacteria were subjected to pectinase enzyme assay in triplicate using the method of Oumer & Abate[9]. The isolates were grown in citrus pectin broth and the initial number of inoculum was adjusted to 0.5 McFarland standard. A total of 1.5 mL of culture was then centrifuged at 10,000 rpm for 5 min to extract crude pectinase enzyme. Likewise, the substrate was prepared by adding 0.5% w/v citrus pectin to pH 7.5 0.1 M phosphate buffer. A volume of 100 μL of crude enzyme was added to a test tube containing 900 μL of substrate. Shortly, reagent blank was prepared by adding 100 μL of distilled water to a test tube containing 900 μL of substrate, while a test tube containing only 900 μL of substrate served as the enzyme blank. The test tubes were placed in a water bath at 50 °C for 10 min. Two millimeters of dinitrosalicylic acid reagent (DNS) was then added to the test tubes to terminate the reaction. Meanwhile, 100 μL of crude enzyme was added to the enzyme blank, then all test tubes were again incubated in the water bath at 92 °C for 10 min. Lastly, the tubes were allowed to cool and optical density (OD) at 540 nm was measured using Epoch 2 microplate spectrophotometer (BioTek). Enzyme activity was measured against enzyme blank and reagent blank. The amount of enzyme that catalyzes galacturonic acid at a period of time (μmol·min−1) was determined based on the OD readings.
Hydrolytic enzyme production of pectinolytic bacteria
-
The pectinolytic bacteria isolates were also subjected to other hydrolytic enzyme activities, such as amylase, cellulase, and protease.
The amylase production of the pectinolytic bacteria was screened using the starch medium, containing 0.5 g·L−1 peptone, 3 g·L−1 beef extract, 0.5 g·L−1 NaCl, 10 g·L−1 soluble starch, and 12 g·L−1 agar. Similarly, 4 μl of 24-h broth culture inoculum was spot-plated in triplicate on the starch agar medium and plates were incubated at 30 °C for 48 h. Agar plates were then flooded with Gram’s iodine solution to observe for clearing around colonies.
Cellulase activity was screened using the carboxymethyl cellulose (CMC) agar medium, containing 0.5 g·L−1 KH2PO4, 0.25 g·L−1 MgSO4, 2 g·L−1 carboxymethyl cellulose, 0.2 g·L−1 Congo red, 2 g·L−1 gelatin, and 12 g·L−1 agar. Agar plates were incubated at 30 °C for 24 h and clear zones around colonies were observed.
Protease production was evaluated using the non-fat milk agar medium, containing 3 g·L−1 peptone, 1 g·L−1 yeast extract, 100 mL·L−1 UHT non-fat milk, and 12 g·L−1 agar. Spot plating was used by inoculating 4 μl of 24-h inoculum on milk agar medium in triplicate and incubated at 30 °C for 48 h. Clear zones around the colonies indicate a positive protease activity.
Fermentation of carbohydrates
-
Pectinolytic bacterial isolates were grown on different carbohydrates (i.e., glucose, fructose, galactose, sorbitol, mannitol, sucrose, lactose, and maltose) to screen their ability to ferment sugars and produce acids. We adopted the standard carbohydrate fermentation protocol of Reiner[10] using phenol red carbohydrate broth, containing 10 g·L−1 peptone, 5 g·L−1 NaCl, 1 g·L−1 beef extract, 0.018 g·L−1 phenol red, and 10 g·L−1 carbohydrate. A color change into yellow indicates fermentation of sugars due to the production of acids in the medium.
Abiotic stress tolerance of pectinolytic bacteria
-
The bacterial isolates were subjected to abiotic stress tolerance on varying temperatures (4, 22, 45 °C) , pH (4, 7, 9), salt (3%, 7%, 10% NaCl), and alcohol levels (1%, 3%, 5% ethanol) using a nutrient broth substrate. Turbidity indicates bacterial growth and tolerance to specific conditions.
Molecular identification
-
The genomic DNA of pectinolytic bacteria were extracted using Vivantis GF-1 Bacterial DNA Extraction Kit, following manufacturer's protocol. The quality of DNA was verified in 0.8% agarose gel dissolved in 0.5X TAE buffer using gel electrophoresis and gel documentation system (Vilber Lourmat). The concentration and purity of DNA were quantified using a NanoDrop 2000c UV-Vis Spectrophotometer (Thermo Scientific™).
The extracted genomic DNA was subjected to PCR amplification by targeting the 16S rRNA gene using universal primers 27f (5'-AGAGTTTGATCCTGGCTCAG-3') and 1492r (5'-GGTTACCTTGTTACGACTT-3'). PCR was performed in a 50-uL reaction containing 1X Taq Master Mix (Vivantis), 2 mM MgCl2, 0.2 μM each of 27f and 1492r primers, and 100 ng of DNA template. A final volume of 50 μL was adjusted with sterile nano pure water. PCR reactions were performed using MiniAmp Plus thermal cycler (Applied Biosystems™) with the following conditions: initial denaturation step at 95 °C for 5 min, followed by 30 cycles of 94 °C for 30 s, annealing at 55 °C for 30 s, and 72 °C for 1 min, with a final extension step of 72 °C for 10 min. The PCR products were verified through gel electrophoresis using 1.2% agarose gel (stained with GelRed®) under 100 V for 35 min and sent for sequencing.
The 16S rRNA gene sequences obtained were compared with the NCBI database through BLASTn searches. The closely related sequences with ≥ 98% similarity were downloaded as a FASTA file, aligned using Muscle software with default settings, and constructed to a phylogenetic tree by Neighbor-Joining algorithm in MEGA software[11,12]. The identity of the bacteria was verified based on the clustering of the target sequence with the closest annotated sequence in the tree.
-
During fermentation, the pH level decreases as the process progresses. For this 24-h time point fermentation, the lowest pH recorded was 4.024 (Table 1). The temperature remained constant at 26 °C in different time points since the set-up is controlled. For a 48-h fermentation, the maximum rate of total aerobic bacteria (7.05 × 107 CFU ml−1) was found at 24-h. A rapid rise of the bacterial population, from 6.95 × 105 to 3.21 × 107 CFU ml−1, was observed within the first 12 h of the fermentation process (Table 1).
Table 1. Changes in pH and culturable population of total aerobic bacteria in 12-h interval time points over 48-h Arabica coffee fermentation.
Environmental
factorsFermentation time 0-h 12-h 24-h 36-h 48-h pH 6.518 5.083 4.024 4.633 4.451 Total aerobic
bacteria
(CFU ml−1)*6.95 × 105 3.21 × 107 7.05 × 107 6.40 × 107 3.60 × 107 * CFU (Colony forming unit) after 24 h incubation. A total of 35 bacterial colonies were randomly selected based on their colony morphological differences. The pure cultures exhibited a wide range of colony characteristics, including margin, shape, elevation, and color (data not shown). The pure cultures isolated in the present study are all Gram-negative and rod-shaped bacteria.
Hydrolytic enzyme activities of pectinolytic bacteria
-
The study successfully screened five pectinolytic bacteria based on their pectinase activity on the citrus pectin agar medium (Fig. 1a). The pectin solubilization index ranges from 3.75 to 5.33 and the pectinase activity ranges from 1.222 to 1.268 μmol·min−1 by which isolates P5B3.4 and P3TA.1 exhibited the highest enzymatic index and activity (Table 2).
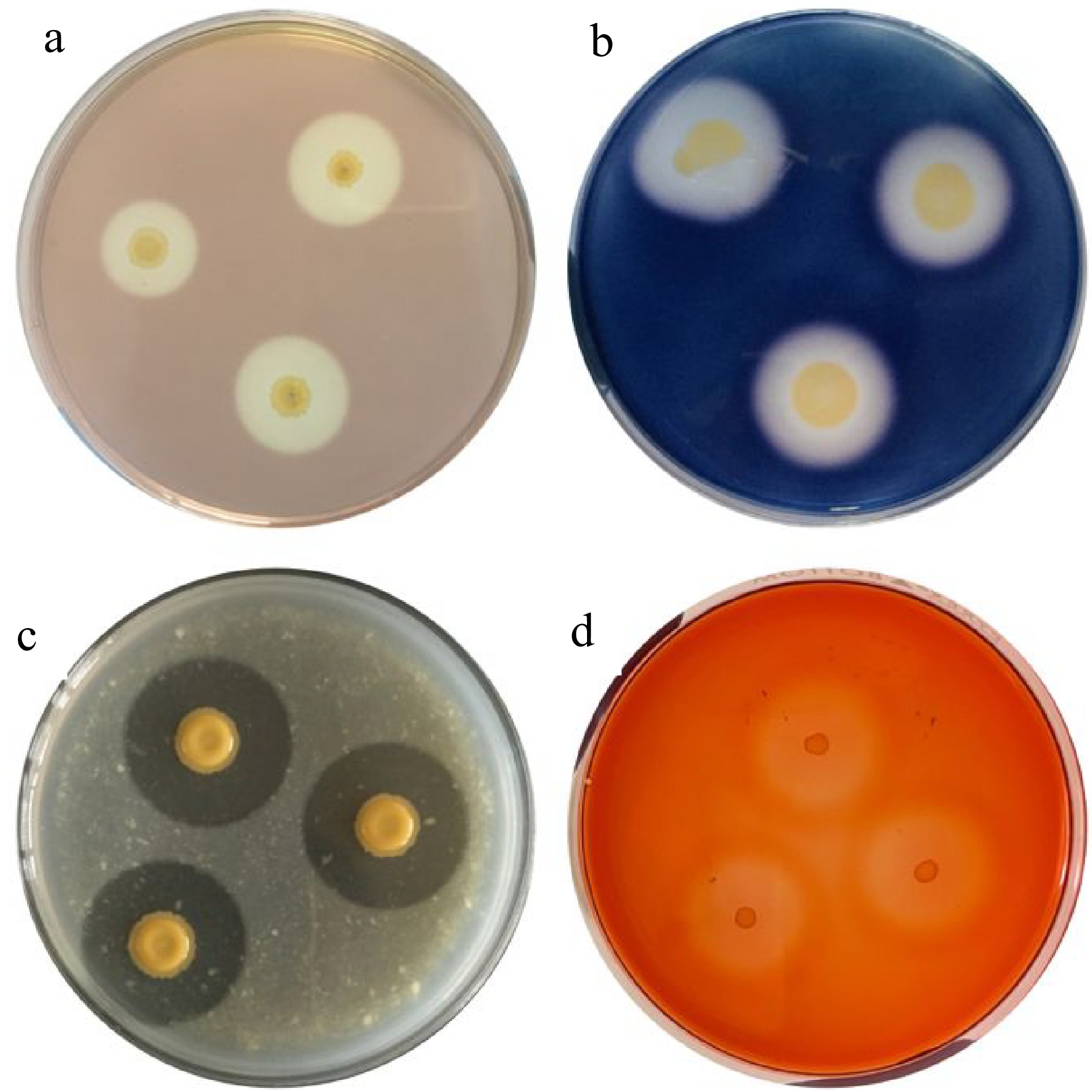
Figure 1.
Enzymatic activities of bacteria from coffee fermentation: (a) pectinase, (b) amylase, (c) protease, and (d) cellulase.
Table 2. Pectinase activity of bacterial isolates obtained from Arabica coffee fermentation.
Isolate Solubilization index* Enzymatic activity (μmol·min−1)* P5TC.3 5.16 ± 0.05 1.257 ± 0.038 P3TD.5 4.13 ± 0.00 1.246 ± 0.019 P5TA.4 3.75 ± 0.11 1.222 ± 0.012 P5B3.4 5.33 ± 0.00 1.263 ± 0.018 P3TA.1 5.33 ± 0.00 1.268 ± 0.010 * Values are mean ± SD in triplicate. In terms of their other hydrolytic enzyme production, all of these pectinolytic bacteria were able to produce amylase, protease, and cellulase (Table 3) based on the solubilization activities (clearing around the colonies) on the starch agar (Fig. 1b), milk agar (Fig. 1c), and CMC Congo red agar (Fig. 1d) plates, respectively.
Table 3. Characteristics of pectinolytic bacteria from fermenting arabica beans.
Characteristic P5TC.3 P5TA.4 P5B3.4 P3TA.1 P3TD.5 Gram reaction − − − − − Shape Rods Rods Rods Rods Rods Hydrolytic enzyme activity Pectinase + + + + + Cellulase + + + + + Amylase + + + + + Protease + + + + + Fermentation of (acid production from) carbohydrates Glucose + + + + + Fructose + + + + + Galactose − + − − + Sorbitol − + − − + Mannitol − + − − + Sucrose + + + + + Maltose + + + + + Lactose − + − − + Growth at different temperatures 4 °C +* + + + +* 22 °C + + + + + 45 °C − + − − + Growth at different pH conditions 4 + + + + + 7 + + + + + 9 − + − + + Growth at different NaCl concentrations 3% + + + + + 7% − + − − + 10% − + − − − Growth at different alcohol levels 1% + + + + + 3% + + + + + 5% − − − − − Legend: (+) positive; (+*) weak growth; (−) negative Fermentation of different sugars
-
Table 3 shows that all of the bacterial isolates are capable of fermenting multiple sugars, especially P5TA.4 and P3TD.5 that ferment all of the eight sugars tested. The rest only ferment glucose, fructose, sucrose and maltose.
Tolerance to abiotic stress
-
All pectinolytic bacteria relatively exerted tolerance to different temperatures, pH, salt, and alcohol concentrations. Bacterial isolates grow poorly and slowly at 4 °C, whereas P5TA.4 and P3TD.5 tolerated 45 °C. For acid tolerance, all isolates are capable of thriving in a pH 4 medium and mostly tolerate the pH 9. Among them, isolate P3TD.5 tolerate up to 7% NaCl while P5TA.4 tolerate up to 10% NaCl concentration. Moreover, all isolates are capable of growing up to 3% alcohol (Table 3).
Molecular identity of pectinolytic bacteria
-
Based on the 16S rRNA gene analysis with ≥ 98% similarity, the pectinolytic bacteria are all Gram negative and rod-shaped cells identified as Chryseobacterium bernardetii (98.25%, 98.52%), Chryseobacterium indologenes (99%), Enterobacter hormaechei (99%), and Klebsiella variicola (99%) (Table 4).
Table 4. Molecular identities of pectinolytic bacteria obtained from coffee fermentation.
Isolate Closest neighbor (type strain) Similarity Identity Accession no. P5TC.3 Chryseobacterium indologenes strain WZE87 99% Chryseobacterium indologenes HQ848390.1 P3TD.5 Klebsiella variicola strain DX120E 99% Klebsiella variicola CP009274.2 P5TA.4 Enterobacter hormaechei strain RPK2 99% Enterobacter hormaechei KX980424.1 P5B3.4 Chryseobacterium bernardetii strain G229 98.52% Chryseobacterium bernardetii JX100816.1 P3TA.1 Chryseobacterium bernardetii strain G229 98.25% Chryseobacterium bernardetii JX100816.1 -
Microorganisms like bacteria play a huge role in fermentation processes[13]. A study suggests that the microbiome profiling in coffee fermentation is dominated by lactic acid bacteria (LAB) and acetic acid bacteria (AAB) after 6 h and the acid tolerant bacteria remains until the end of the process[14]. The organic compounds and acids released by LAB, AAB and other fermenting microbes can accumulate in the fermentation setup, creating a more acidic environment[15]. Our study recorded the lowest pH of 4.024 at 24 h fermentation time where the highest population of aerobic bacteria was also recorded (Table 1). Similarly, it was reported that the population of total aerobic bacteria really increased as the fermentation progressed[15,16]. Meanwhile, the present study isolated rod-shaped and Gram-negative bacteria, which corroborated with the study of Pregolini et al.[14], suggesting that members of Enterobacteriaceae are present in higher frequencies even at the beginning of the fermentation.
The beans of a coffee cherry are surrounded by different layers, including the sticky polysaccharide layer called pectin[5]. Pectinase production is important to easily digest the sticky pectin substances in the mucilage by breaking the α-1,4-glycosidic bonds in pectin[17]. The production of pectinolytic enzymes and formation of alcohols and acids (butyric acetic, lactic, and other long-chained carboxylic acids) are associated with microbes such as pectinolytic bacteria[5]. Our study successfully screened and identified pectinolytic bacteria obtained during Arabica fermentation, showing their potential in accelerating mucilage degradation. Besides, for complete pectin degradation in coffee mucilage, three enzymes are involved, including polygalacturonase, pectin lyase, and pectin methylesterase[7]. Thus, the pectinolytic trait of certain species of bacteria poses a vital role as potential starter culture for coffee fermentation. On the other hand, other hydrolytic enzymes such as amylase[18], cellulase[19], and protease[20] are commonly produced by a broad range of microorganisms, possessing characteristics of biotechnological interest and industrial applications. In the present study, multiple hydrolytic enzyme production of these pectinolytic bacteria revealed promising potential as starter cultures in coffee fermentation. Also, these bacteria were able to ferment different sugars and produced acids, indicating their ability to perform fermentation (Table 3).
Coffee from the wet fermentation processing method is more acidic than the others due to lowering of pH by bacteria during wet fermentation[21], thus tolerance to this stress helps starter culture to perform optimum process during fermentation. The tolerance of these pectinolytic bacteria in a wide range of temperature, pH level, high salt and alcohol contents contribute to their roles as potential starter cultures (Table 3). The activity of starter cultures on fermentation impacts bioactivity, stability, antimicrobial activity and toxicity of the finished product[22], especially when exposed to abiotic stresses. The ability of these bacteria to develop resistance or tolerance to multiple stresses can be attributed to their ability to maintain pH homeostasis, cell membrane integrity and fluidity, metabolic regulation, and macromolecule repair[23].
The prokaryotic 16S rRNA gene (~1,500 bp) is composed of variable regions interspersed between conserved regions[24]. This gene has become the gold standard to identify bacteria and establish taxonomic relationships between prokaryotes, with 98.65% similarity as the threshold for delineating species[25]. Our study has isolated species of Gram-negative, rod-shaped bacteria that were molecular identified as Chryseobacterium bernardetii, Chryseobacterium indologenes, Enterobacter hormaechei, and Klebsiella variicola. The coffee fermentation microflora are rich and mainly constituted of aerobic Gram-negative, rod-shaped bacteria[26]. The genera Chryseobacterium, Enterobacter, and Klebsiella were found to be part of the core coffee microbiota in different plant compartments (i.e., rhizosphere, episphere, and endosphere)[27], and their pectinolytic activities were previously reported[28−30]. Moreover, the genera Enterobacter and Klebsiella are prevalent at the beginning of fermentation process[31], but to our knowledge there was no report of Chryseobacterium species in coffee fermentation. Nevertheless, fermentation affects the metabolic activities of these natural microbiota, which predominantly grow and uniquely impact the coffee quality[30].
-
The study isolated indigenous pectinolytic bacteria that show promising potential for starter cultures in coffee fermentation. Aside from pectinase, these bacteria also produce other hydrolytic enzymes (i.e., amylase, protease, and cellulase) and are capable of fermenting different types of sugars. Their tolerance to a wide range of abiotic stresses pose an additional promising role as potential starter cultures. Molecular identification confirmed that they belong to the genera Chryseobacterium, Enterobacter, and Klebsiella, which are found to be part of coffee microbiota. These findings suggest the potential use of pectinolytic bacteria as starter cultures in coffee fermentation.
-
The authors confirm contribution to the paper as follows: study conception and design: Cortes A, Baltazar M; material preparation and data collection: Cortes A, Baldomero JR; analysis and interpretation of results: Cortes A, Baldomero JR; draft manuscript preparation: Cortes A; review and editing: Baltazar M; fund acquisition: Baltazar M, Cortes A. All authors read and approved the final manuscript.
-
All data generated and analyzed are included in this paper.
The study was funded by US Department of Agriculture through ACDI-VOCA under the PhilCAFE In-Kind Grant project (Grant No: 718948656). We are grateful to the Equilibrium Intertrade Corporation for providing Arabica coffee cherry samples and assistance during collection as well as to Mr. Maowel Villanueva for the technical assistance during sample collection and processing.
-
The authors declare that they have no conflict of interest.
- Copyright: © 2024 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Cortes AD, Baldomero JRN, Baltazar MD. 2024. Molecular identification of indigenous pectinolytic bacteria characterized for starter culture in coffee fermentation. Beverage Plant Research 4: e026 doi: 10.48130/bpr-0024-0015
Molecular identification of indigenous pectinolytic bacteria characterized for starter culture in coffee fermentation
- Received: 30 December 2023
- Revised: 16 February 2024
- Accepted: 11 March 2024
- Published online: 10 July 2024
Abstract: From cherries to green beans, coffee undergoes a post-harvest fermentation process. The quality of coffee is influenced by the origin and microbiological activities that drive coffee fermentation, particularly pectin hydrolysis. Coffee-associated pectinolytic microorganisms have been isolated and characterized to explore their potential as starter cultures for coffee fermentation. This study characterizes the indigenous pectinolytic bacteria for starter cultures, which were isolated during the wet fermentation of Coffea arabica cherries. A total of five indigenous bacteria had the ability to produce pectinase enzymes with solubilization index ranging 3.75−5.33 and enzymatic activity ranging 1.22−1.268 μmol min−1. Interestingly, these bacteria showed amylase, cellulase, and protease activity in addition to pectinase. All of them are capable of fermenting multiple sugars and releasing acids. Moreover, they tolerate a wide range of fermentation stress (i.e., temperature, pH, salt, and alcohol). Based on the 16S rRNA gene sequencing, they were designated as Chryseobacterium bernardetii (P5B3.4 and P3TA.1), Chryseobacterium indologenes (P5TC.3), Enterobacter hormaechei (P5TA.4), and Klebsiella variicola (P3TD.5). The genera of these pectinolytic bacterial species are part of coffee microbiota and found to be associated with coffee cherries. Thus, they pose potential use for starter culture in coffee fermentation in the Philippines.
-
Key words:
- Arabica coffee /
- Fermentation /
- Pectinolytic bacteria /
- Starter culture













