-
Black rice is a rare variety of rice with a broad geographical distribution[1]. The content of trace elements manganese and zinc is 1−3 times higher than that of ordinary rice. Moreover, it encompasses distinctive components such as vitamin C, chlorophyll, and anthocyanins. As a result, black rice possesses a higher nutritional value compared to regular rice. Recent research has demonstrated that black rice possesses antioxidant, anti-inflammatory, and anti-tumor properties, as well as the ability to improve type 2 diabetes[2,3]. Additionally, it has been shown to prevent the occurrence of cardiovascular and cerebrovascular diseases, along with exhibiting other distinct biological activities[4]. The physiological functions of these compounds are likely attributable to their anthocyanin content[5]. However, their stability is relatively low and they are susceptible to degradation under external conditions. Anthocyanins in black rice are primarily comprised of centaurin-3-O-glucoside (C3G), which accounts for approximately 88% of the total anthocyanin content[6]. To improve the stability of anthocyanins, the structural modification and manipulation of environmental conditions have been used in recent studies[7−9]. The specific structural methods encompass copolymerization, acylation, and biosynthesis. Environmental approaches involve liposomes, microencapsulation, and encapsulation of nanoparticles[10]. However, these techniques are prone to certain limitations. For instance, acylation may introduce potentially detrimental residues, while, encapsulation has the potential to decrease water solubility.
During the food production process, anthocyanins can interact with a variety of proteins. It is also essential for anthocyanins to form complexes with carrier proteins to facilitate normal metabolism, transportation, and absorption in vivo[11]. The interaction can be either non-covalent or covalent, depending on the chemical structure of the reactants and the reaction conditions[12], which further influences the conformation of carrier proteins[13]. Consequently, the investigation of exploration between anthocyanins and proteins is indispensable for understanding the in vivo transport and metabolism of anthocyanins at a molecular level. Currently, protein binding has been demonstrated to be an effective approach for the stabilization of anthocyanins[14]. The enhancement of stability is related to the molecular structure of the complex. At present, the stability of anthocyanins can be improved by changing the structure of the complex, but the mechanism of action is not yet clear. Bovine serum albumin (BSA), as one of the predominant transport proteins in plasma, plays a crucial role in maintaining the stability of plasma colloid osmotic pressure, facilitating material exchange with interstitial fluid[15]. It also possesses a distinct hydrophobic cavity that serves as a binding site for anthocyanins, facilitating their interaction[16]. Consequently, it is of great significance to investigate the binding mechanism between BSA and anthocyanins.
External static magnetic fields (SMF) have been approved to affect chemical or biological molecular interaction via regulating molecular binding[17]. From a microscopic perspective, all the molecules are composed of atoms. When the number of protons or neutrons is odd, the nucleus becomes a magnetic nucleus and the spin of the magnetic nucleus forms a current loop. It results in the generation of a magnetization vector with specific magnitude and direction, as depicted in Fig. 1. In solution systems, the SMF induces molecular binding among proteins, small molecules, and solvents[18]. The effects of SMF on proteins include the changes of secondary bonds, peptide bonds, and distribution of electrons and molecules[19]. For example, the spatial structure of proteins is primarily maintained by secondary bonds. SMF can lead to the exposure of a distinct number of tryptophan groups, internal tyrosine groups, and aliphatic groups on the protein surface by affecting secondary bonds, such as the disruption of certain disulfide bonds[20]. Secondly, the diamagnetic anisotropy of proteins is contributed by peptide bonds, such as the α-helix structure. Thirdly, the SMF induces alterations in the distribution of electrons and molecules, resulting in the polarization and displacement of atoms and molecules. This subsequently leads to modifications in electron transition probability, dipole moment transition, and molecular vibration state, while maintaining the atomic and molecular composition unchanged[19]. These effects may result in the formation of protein clusters in SMF, commonly denominated as magnetic domains, which significantly enhance the efficiency of protein binding to small molecules. In addition, the effects of the SMF on small molecules mainly encompass molecular distortions, increased interactions, and changes in bond angles. The physico-chemical properties of the reaction solution can be affected by the SMF. Firstly, the viscosity and surface tension of the solution could be affected by the changes in molecular interactions under the inducement of SMF. Secondly, SMF causes the changes in the hydrated ion layer and the hydrated water structure which further influences the water conductivity[20]. For polar compounds with high diamagnetism, the SMF have distinct advantages for inducing intermolecular binding. It can transfer energy to the microstructure of matter without direct contact, which is attributed to the influence of SMF on the mutual transformation of electron triplets and singlets of free radicals. Due to the reason of energy, free radicals in triplets are less prone to Gemini recombination[21]. Moreover, SMF have the capability to alter bond arrangement and orientation. Thus, it could provide superior control over microstructure control through adjusting the direction and intensity of the magnetic field[22].
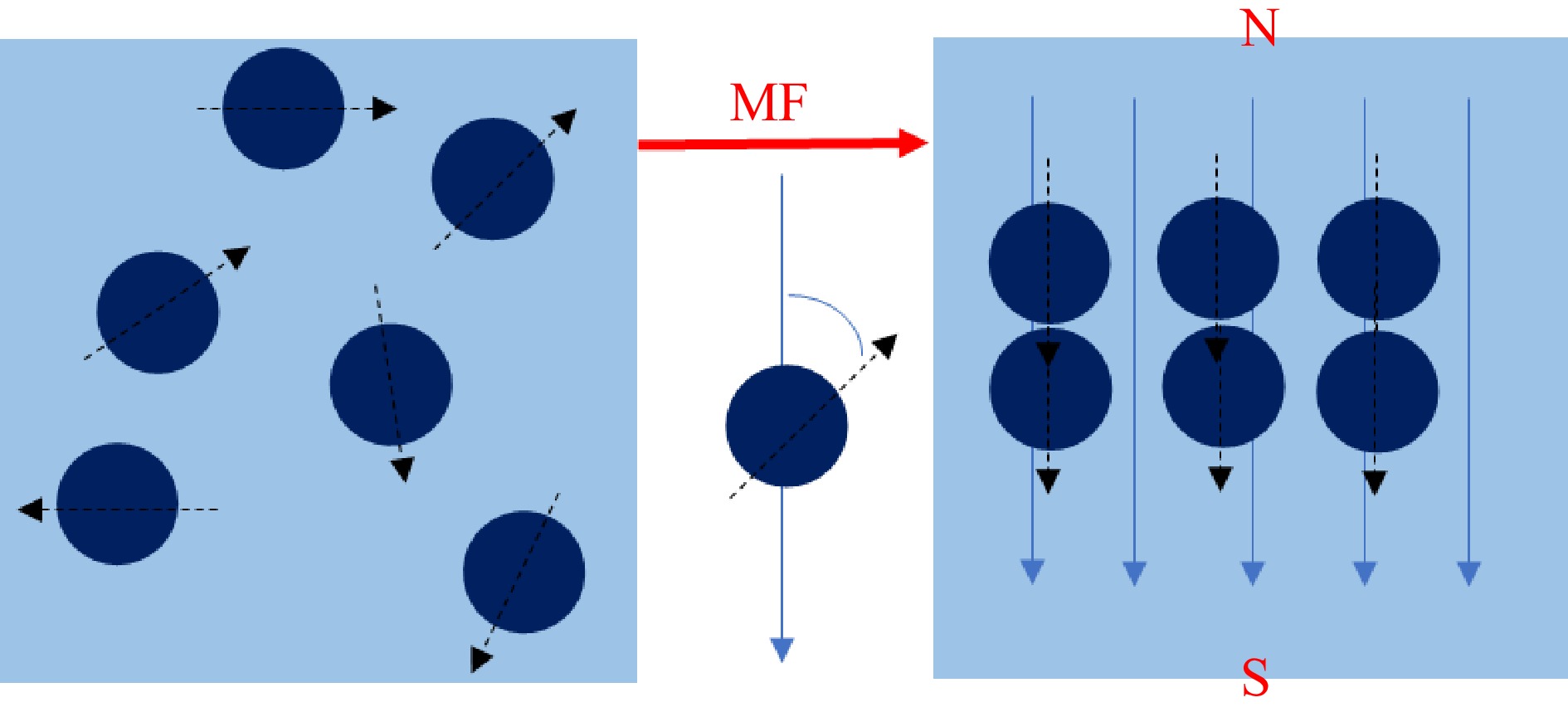
Figure 1.
The spin of a magnetic nucleus produces a magnetization vector (MF: magnetic field, N: magnetic north, S: magnetic south).
In previous experiments conducted by the research group, the SMF had an impact on stability. In this paper, the formation and structural changes of composites during magnetic processing were further explored, and delve into the mechanism of the influence of SMF on the structure of composites. Currently, while the interactions between proteins and small molecules have been documented, the impact of SMF induction on the binding mode between BAS and C3G remains inadequately explored. Therefore, the related mechanism would be revealed by the analysis of secondary structure, functional groups, dipole moment, unit cell size, and microstructure of the complex.
-
BSA ( purity ≥ 97%, GENVIEW), C3G (purity ≥ 98%, Vicky Biotechnology Co., Ltd), potassium bromide (spectrally pure, Tianjin Damao), trimethylol aminomethane (Tris, purity ≥ 99%, Amresco), HCl (Tianjin Damao), anhydrous ethanol (Tianjin Damao), NaCl (Tianjin Damao). All other reagents were domestic analytical pure. The water used in the experiment was tertiary ultra-clean water.
Reagent preparation
-
A Tris-HCl buffer (0.05 mol/L, pH 7.4) of 0.10 mol/L NaCl was prepared to maintain the ionic strength and pH of the solution. A BSA solution (1 × 10−6 mol/L) was prepared with the Tris-HCl buffer and stored at 4 °C for later use. A stock solution of C3G (3 × 10−3 mol/L) was prepared in anhydrous ethanol and stored at 4 °C for later use.
Molecular docking simulation
-
Schrodinger molecular docking software was used to predict the molecular binding conformation of BSA and C3G[23]. First, protein macromolecular file was prepared. In File-Get PDB, the molecular file of BSA was loaded through the functions provided by maestro. The PDB ID was entered as 4F5S and downloaded. The Protein Preparation Wizard module was selected, and the Fill in missing side chains using the Prime option checked under the Import and Process processing box. The conserved water molecules were retained and charged. The hydrogen bond network of amino acid residues were optimized and the energy was minimized. After running, the prepared protein molecular file was obtained. Second, the file of the ligand small molecule was prepared. The CAS number 7084-24-4 of C3G was searched in Pubchem, the 2D structure file (sdf type) downloaded, and then the downloaded file uploaded into maestro. The small molecule file in the LigPrep module was selected, OPLS3e chosen in the Force field, and 'Generate possible states at target pH' chosen when setting the ionization state: 7.0+/−0.5, the following were checked: Epik, Desalt, Generate tautomers, Retain specified chiralities (vary other chiral centers), the 'Generate at most' was set to 32 per ligand, the format set and then perform the operation. After running, the prepared ligand molecular file was obtained. Third, a SiteMap was run on a protein molecule to look for pockets of activity. The option for the whole macromolecule was set in the SiteMap module. The precision setting requires at least 15 site points per reported site, Report up to 5 sites (site-point groupings), and Crop site maps at 4 Atoms come from nearest site point and run. Fourth, Receptor pocket files were generated under the Receptor Grid Generation module and molecular Docking performed under the Ligand Docking module.
Magnetic processing complex
-
The SMF required for the experiment was provided by a 100 mm × 100 mm × 20 mm Ndfeb magnet, which was purchased from Shanyang District, Jiaozuo City, Xin Heng strong magnetic hardware store (China). The two Ndfeb magnets were fixed in parallel, and the SMF strength changed by changing the distance between the two magnets. The required magnetic induction intensity (50 mT-200 mT) was determined by the Tesla meter. BSA solution (1 × 10−6 mol/L) and C3G solution (3 × 10−3 mol/L) were mixed with a volume ratio of 1:1. The samples were then treated in a SMF of 50, 100, 150, and 200 mT for 4 h, respectively.
Circular dichroism chromatography
-
The secondary structure changes of the samples were determined by circular dichroism. The experimental instrument is a circular dichrometer (Chirascan V100, applied photophysics, UK). The response time was 0.5 s, the scanning rate was 100 nm/min, the slit width was 2 nm, and the step size was 1 nm. Then, the circular dichrograms of each sample were collected. The secondary structure of polypeptide was calculated, and the content and proportion of each sample were obtained.
Fourier infrared spectroscopy
-
The samples were ground to less than 200 mesh and dried in a drying oven for 4 h until no clumping appeared. Appropriate amount of powders (1−2 mg) were ground with 200 mg potassium bromide, mixed, and pressed into a tablet. The samples with the treatments of 0 and 200 mT were tested, and the absorption spectra were determined by a Fourier infrared spectrometer. The experimental instrument used was a Fourier transform infrared spectrometer (IRAffinity-1, Shimadzu, Japan). The parameters included a wall-number range of 4,000 to 400 cm−1, 64 scans with an average resolution of 4 cm−1, and an ambient temperature of 25 °C.
X-ray wide-angle diffraction
-
The experimental instrument was a X-ray diffractometer (ADVANCE, Brook, Germany). Three grams of the lyophilized sample was ground to a particle size of 40 μm and pressed into tablets. The parameters were as follows: the emission current was 25 mA; the working temperature was 25 °C. The time/step length: 1 s/step length; Interval: 2θ = 4−40°; Scan step size: 0.01. The diffraction peak of the results was smoothed, the back and bottom were subtracted, and the instrument was widened.
Scanning electron microscope
-
The experimental instrument was a Field emission scanning electron microscope (SIGMA500, Zeiss, Germany). Freeze-dried samples were uniformly fixed on the glue-attached electron microscope injection stage and sprayed gold under vacuum conditions. They were then fixed on the stage to adjust the best field of view and magnification for observation.
Statistical analysis
-
Each experiment was repeated three times. The experimental data were processed and analyzed by Excel and Origin 2022b, and the correlation analysis was performed by SPSS 26.0, and the significance level was p < 0.05.
-
Molecular docking is a computer simulation program used to predict the conformation of receptor-ligand complexes[24]. Molecular docking technology can simulate the binding between C3G and BSA, which helps to understand the ligand-receptor interaction better and further verify the experimental conclusions. It has been reported that the degree of hydroxylation on the B-ring of anthocyanins determines the hue and color stability of anthocyanins[25]. The antioxidant capacity of anthocyanins is associated with the number of hydroxyl groups in the B ring. The hydroxyl group at position 4 of the B ring is the most active group[26]. Figure 2 showed that C3G was mainly bound to the II and III domains of BSA, and six amino acid residues docked with C3G molecule. ASP108, LYS114, ARG144, ARG185, and LEU454 interacted with C3G through hydrogen bonding, resulting in the loss of hydrogen donor which further limited its antioxidant properties. Meanwhile, ARG458 was docked to C3G through cation-π interaction. Hence, the hydrogen bond and cation-π interaction are the main force types in the binding process of C3G to BSA. Recent studies have revealed that the predominant binding mechanism between the two entities is non-covalent binding[27,28], which aligns with our initial prediction. The calculated minimum binding energy of the molecular model was determined to be −7.291 kcal/mol (30.52 kJ/mol). These findings suggest that application of a magnetic field may influence the cation-π interaction and subsequently alter the binding conformation of the two entities[29].
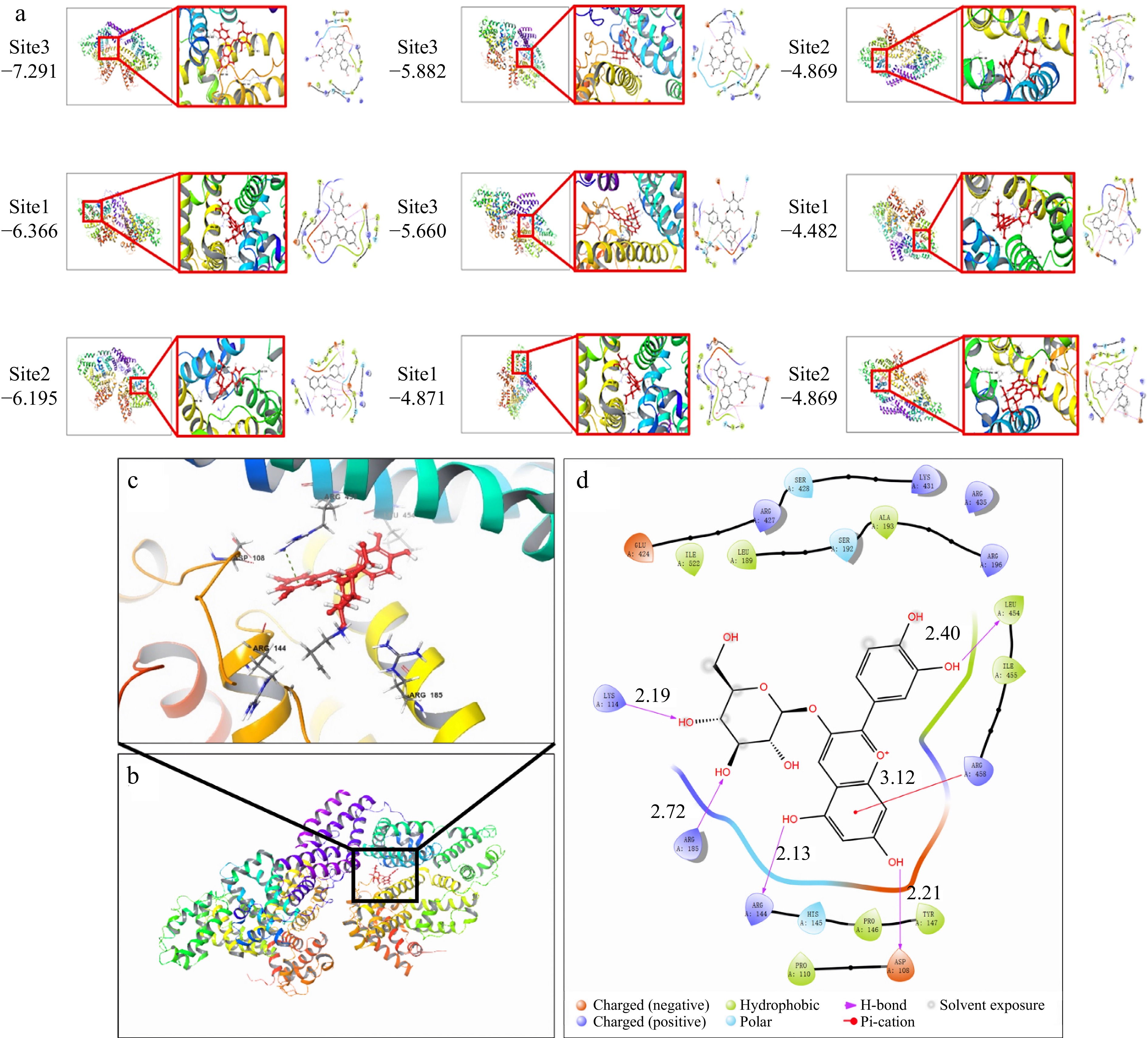
Figure 2.
Molecular docking of simulated BSA-C3G conjugates, (a) nine simulation results, (b) BSA-C3G conjugate model, (c) BSA-C3G binding site detail diagram, (d) main force type.
Effect of SMF on the secondary structure of BSA-C3G composite
-
Circular dichroism (CD) spectroscopy is employed to further investigate the impact of SMF treatment on the binding of C3G-BSA complex (Fig. 3). The complexes subjected to different magnetic fields exhibited two distinct negative absorption peaks at 208 and 221 nm, respectively, indicating the characteristic α-helix structure in the secondary conformation[30,31]. As the intensity of SMF increased, distinct changes were exhibited in the CD of BSA. It suggested that the magnetic field disrupted the protein structure, causing BSA more susceptible to binding with anthocyanins[32]. The experimental results demonstrate that the alterations in absorbance and secondary structure are reverse reactions. Research has found that sometimes the changes in absorbance are small, while the changes in structure are large[33]. In this experiment, this might be attributed to the magnetic field acting on the composite, causing it to form a special structure that influences the absorption of light. As shown in Table 1, an escalation in SMF induction intensity led to a decrease in α-helix content from 32.6% to 23.4%, an increase in β-fold content from 5.5% to 39.6%, a decrease in β-angle content from 22.1% to 8.4%, and a reduction in random coil content from 39.8% to 28.6%. The decrease in α-helix content from 32.6% to 23.4% can be attributed to several factors. For example, the C=O bond of the amide group is capable of forming hydrogen bonds with other functional groups, thereby contributing to the overall secondary structure of the protein complex. The typical α-helix structure is a helical conformation constituted by a hydrogen bond between the C=O of the amino acid at position X and the N-H of the amino acid at position X-4 in the peptide backbone[34]. Magnetic fields can effectively facilitate the transition of hydrogen bonds from disordered to ordered states[35]. In a randomly coiled polypeptide chain, the dipole moment of a single backbone amide group is oriented randomly, resulting in neighboring helices neutralizing each other's dipoles in opposite directions. In α-helices, the hydrogen bond neutralizes their horizontal dipole moments, and the vertical dipole moments point in the same direction[36]. The dipoles of the individual peptides within the helix were combined to form large dipoles. Meanwhile, the amino-terminal pole of the helix becomes positive and the carboxyl-terminal pole becomes negative. Therefore, the charge distribution is asymmetric in the high helical structure of BSA, which reduces the alpha-helical content under the applied magnetic field. The change in the secondary structure of BSA, suggested that C3G bound with amino acids on the main chain of BSA and destroyed the hydrogen bond network.
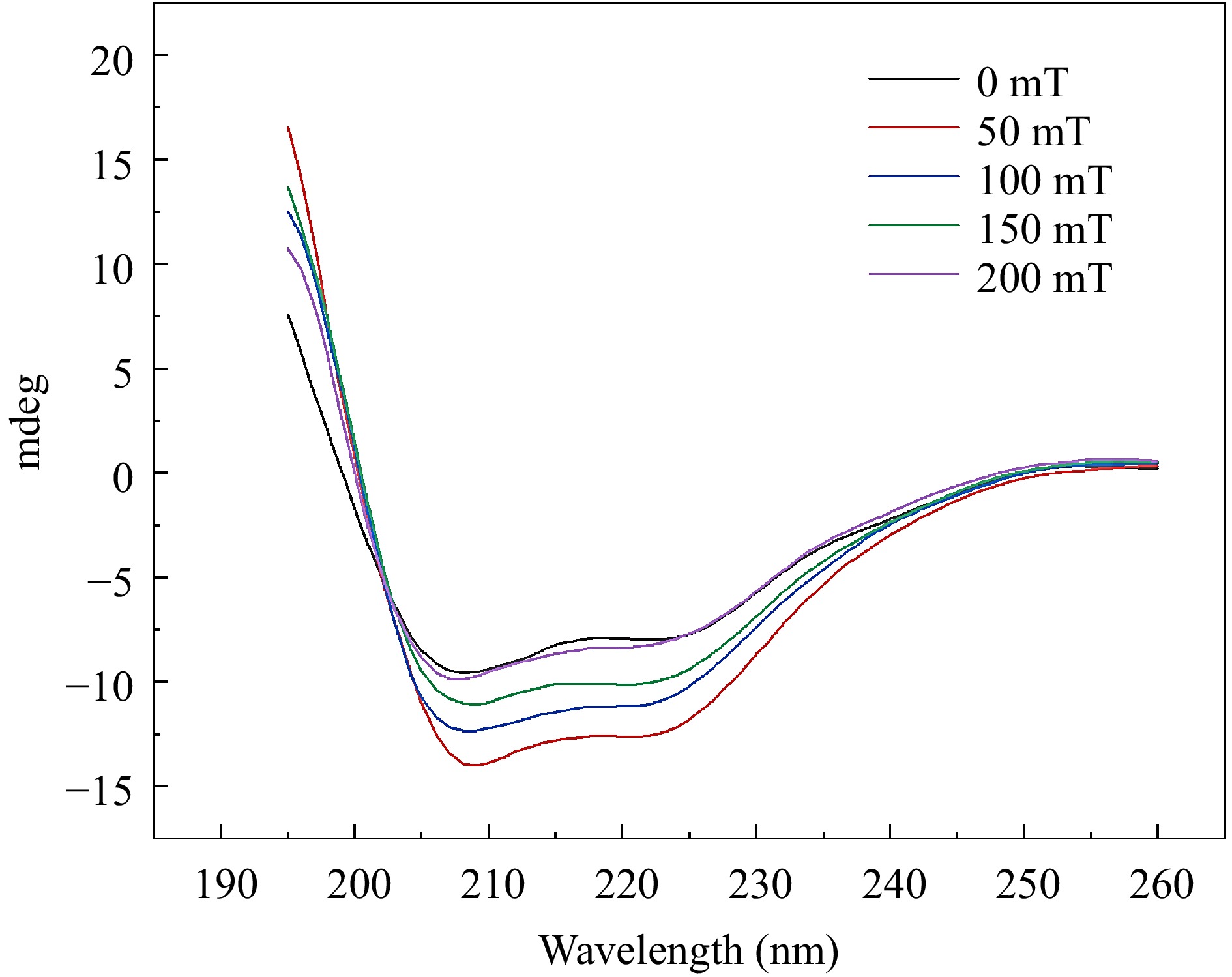
Figure 3.
Circular dichroism spectra of BSA-C3G conjugates at different magnetic sensing intensities.
Table 1. Changes in secondary structure content of BSA-C3G conjugates at different magnetic sensing strengths.
Magnetic intensity α-helix β-sheet β-turn Random coil 0 mT 32.6% 5.5% 22.1% 39.8% 50 mT 29.3% 23.7% 16.9% 30.1% 100 mT 25.0% 30.9% 12.1% 31.9% 150 mT 24.3% 35.8% 10.7% 29.2% 200 mT 23.4% 39.6% 8.4% 28.6% Effect of SMF on the intermolecular interaction of BSA-C3G composite
-
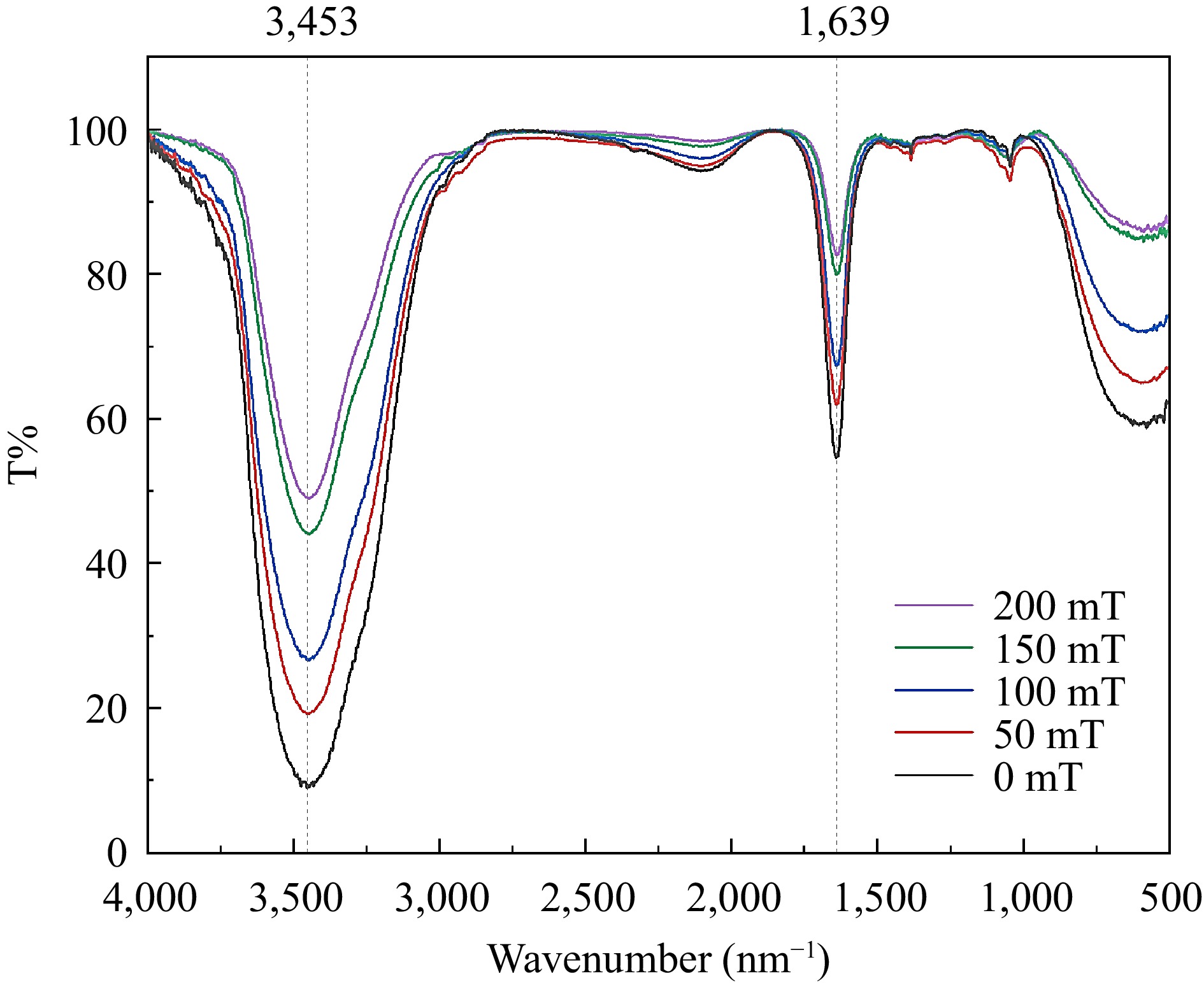
To investigate the potential re-dissociation of the complex into individual BSA and C3G molecules in solution with exposure to an SMF, the solution is analyzed using the infrared spectroscopy[37,38]. Based on the result of Fourier infrared spectroscopy, no separation of the complex after binding was observed (Fig. 4). Furthermore, the position and number of absorption peaks in the complexes different magnetic induction treatments remained unchanged. It was indicated that the chemical bond between BSA and C3G was not influenced by the SMF[39]. However, the transmittance underwent a distinct change, which could potentially be ascribed to the dipole moment of the conjugate[40]. With the increasing intensity of the SMF, the infrared spectral transmittance of the sample increased. It may be related to the change of the molecular force of the SMF, resulting in the change of the dipole moment of the sample. The dipole moment changed greatly and the transmittance of the absorption peak increased greatly. The greater the electronegativity difference between the two ends of the bond and the greater the polarity, the greater the transmission is observed. It is concluded that the SMF treatment did not change the chemical bond, but also changed the dipole moment of the bond (the distance between different atoms), then changed the binding effect of the two molecules. Thus, the stronger the magnetic induction intensity of the static field resulted in the greater change of the dipole moment.
Effect of SMF on the crystal characteristics of BSA-C3G composite
-
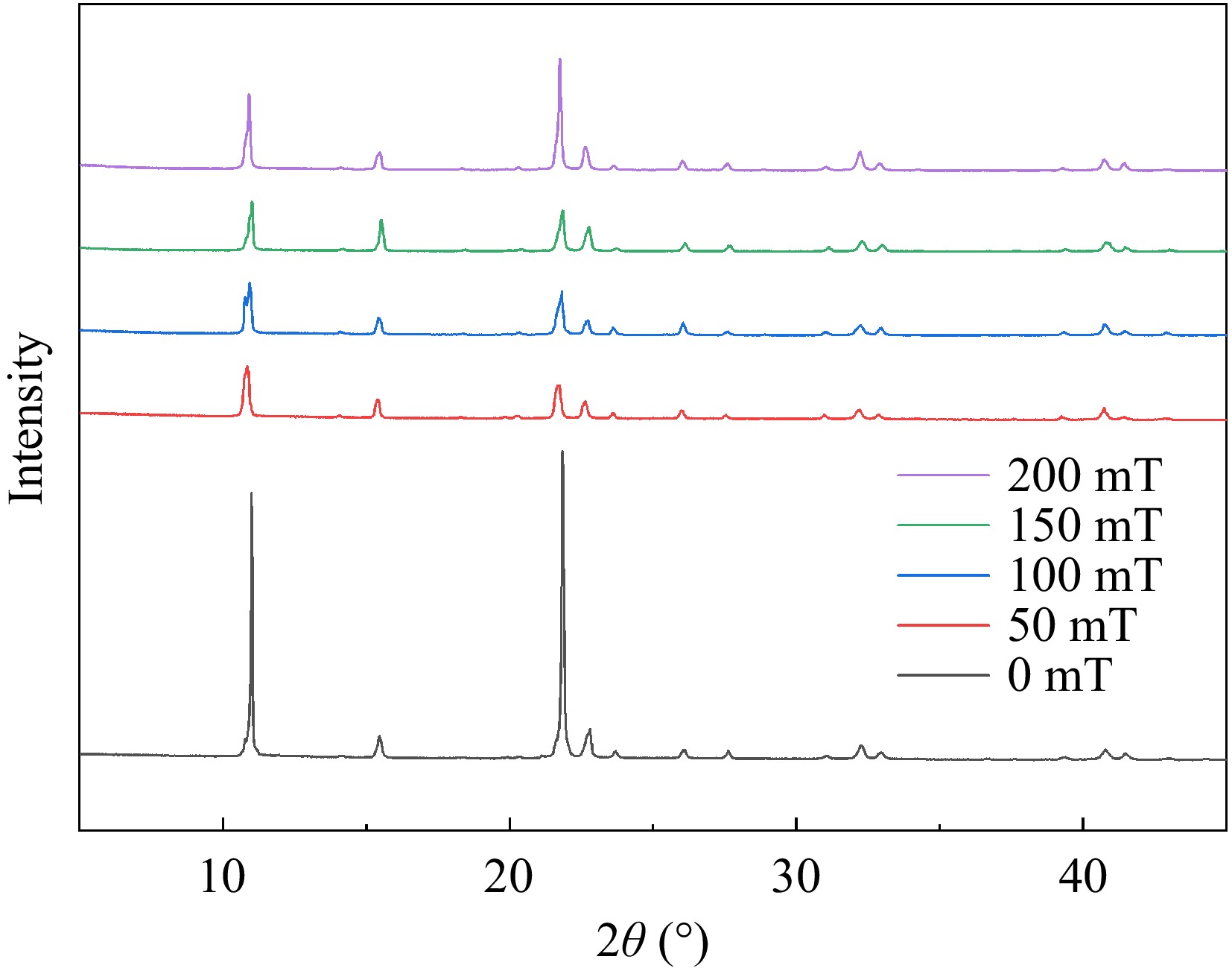
The characteristic diffraction pattern included two primary components: the spatial distribution of diffraction, reflected in the peak position within the characteristic diffraction pattern and the intensity of the characteristic diffraction peaks. The distribution of diffraction peaks is predominantly governed by the size, shape, and orientation of the unit cell. Meanwhile, the intensity is mainly determined by both the type of atoms and their positions within the unit cell. As depicted in Fig. 5, all samples exhibited sharp diffraction peaks indicative of a crystalline structure[41]. The peak heights of samples with SMF treatment were observed to be remarkably high, sharp, and narrow at 11° and 22° compared with the control group. Conversely, the height of the above two peaks decreased clearly in SMF treatment and the peak width was slightly wider than that of 0 mT samples. The alteration in average grain size was also attributed to changes in the peaks at 11° and 22°. According to Diaconu et al.[42], these peaks can be identified as randomly oriented helical structures, suggesting that the influence of SMF on these structures was distinct. This can be attributed to the flexibility of the protein, which allows the restructuring of the structure due to exposure to magnetic forces of different strengths and allowed the two helices to unravel. Generally, maintaining the helical structure mainly depends on hydrogen bonds. After the helix unraveled, more hydrogen bond exposure increased the efficiency of small molecules to bind. Table 2 shows the results of peak position, full width at half maxima (FWHM), and crystaline size. Based on Table 2, the grain size of the samples subjected to the SMF generally decreased. This phenomenon may be attributed to the influence of the SMF on the magnetic dipole moment, with a minimum effect observed at 50 mT and a maximum effect at 200 mT. The treatment of 200 mT showed the highest degree of helix unwinding and the highest efficiency of binding. These results were in accordance with the experimental conclusion of circular binary chromatography.
Table 2. Peak position, FWHM, crystaline size, and average size of conjugates.
Compound Peak position
(2 Theta)FWHM Crystaline
size D (nm)Average D
(nm)0 mT 11.00 0.07 115.30 52.44 15.46 0.20 40.99 21.84 0.11 71.56 22.74 0.25 31.90 23.68 0.19 43.76 27.62 0.15 55.92 32.26 0.24 34.82 32.94 0.20 41.16 40.78 0.23 36.55 50 mT 10.89 0.26 31.14 37.36 21.72 0.27 30.07 23.64 0.16 51.99 26.05 0.18 44.46 31.02 0.20 41.07 32.22 0.30 26.95 32.92 0.20 41.55 40.77 0.27 31.61 100 mT 10.97 0.18 43.81 39.70 15.42 0.16 51.03 21.81 0.23 34.66 22.73 0.24 34.45 27.66 0.19 42.07 31.12 0.19 44.53 32.28 0.26 31.66 32.99 0.23 35.41 150 mT 10.82 0.22 36.57 40.63 15.39 0.17 46.43 20.26 0.17 46.86 21.68 0.23 35.73 22.61 0.21 38.63 25.99 0.18 46.48 30.98 0.18 45.67 32.17 0.25 33.19 40.73 0.23 36.14 200 mT 10.89 0.18 44.21 41.30 21.74 0.15 52.58 26.03 0.20 41.67 32.90 0.24 34.85 40.74 0.26 33.20 Effect of SMF on the binding of C3G to BSA composite
-
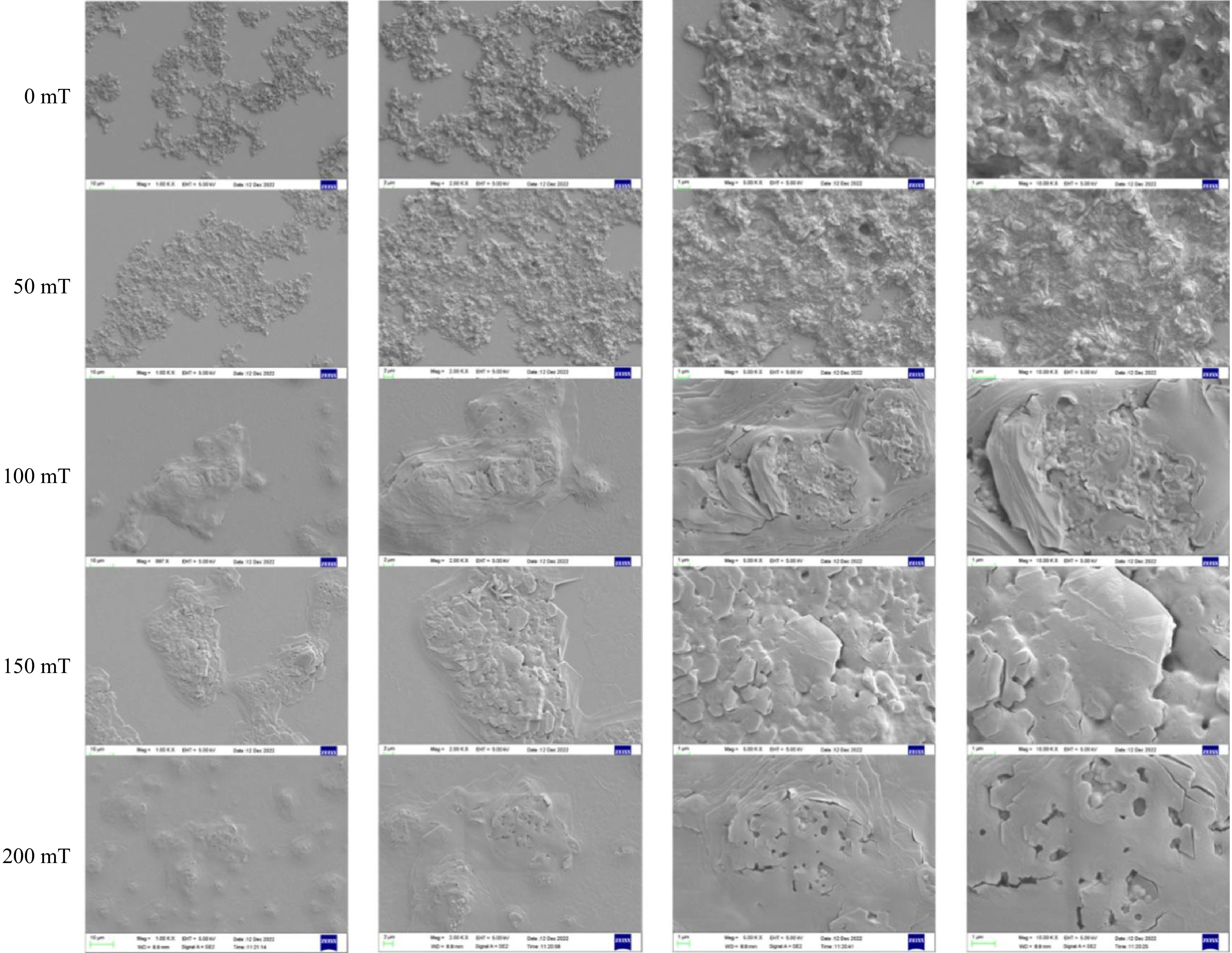
To provide a more intuitive observation of the microstructure of the BSA-C3G complex, scanning electron microscopy was applied[43]. In Fig. 6, the microstructure was observed under different magnetic field strengths and magnified by different observation factors, which shows a more comprehensive and intuitive trend of the composite under a static magnetic field. In the control sample (0 mT), the microscopic surface of the sample revealed aggregated particles in the plane at a magnification of 1 K. In addition to prominent uplift and depression structures, the sample exhibited a rough layer state with fine fracture structures, primarily located at joints within the laminated structure. However, as the magnetic field increased, there was a gradual flattening of the overall structure and an emergence of cavity structures with spherical formations within them.
Scanning electron microscopy revealed the presence of small spherical anthocyanin molecules at magnifications ranging from 1,000 to 10,000 times[44]. After SMF treatment, the previously disordered stacked structure was reorganized into a more regular arrangement. Furthermore, an increase in the magnetic induction intensity of the SMF resulted in a smoother microstructure of the sample.
-
In the present study, the effect and mechanism of SMF on the interaction between BSA and C3G was investigated. The SMF can affect the secondary structure and unit cell size of the complex, induce the interaction between BSA and C3G molecules, and increase the influence of magnetic induction intensity on the complex. This may be due to the higher magnetic induction intensity, the greater the dipole moment between molecules, and the greater the degree of directional rearrangement of complex molecules. These findings provide insights into the mechanism by which SMF interacts with proteins and anthocyanins, provide a basis for SMF to promote their binding, and point out a new possible pathway for improving anthocyanin stability. In future research, the reaction pathway of this complex in vivo can be further explored to investigate its wide targeted reaction process in living organisms.
This research was partially supported by the Natural Science Foundation of Liaoning Province, China (2023-MS-205).
-
The authors confirm contribution to the paper as follows: conceptualization: Li D, Zhang Z; methodology: Zhang Z, Shen Y; data curation: Zhang Z, Xin G; formal analysis and visualization: Zhang Z, Deng W; writing - original draft: Zhang Z; writing-review & editing: Zhang Z, Deng W, Adel Ashour A, Tan H; funding acquisition and supervision: Li D. All authors reviewed the results and approved the final version of the manuscript.
-
All data generated or analyzed during this study are included in this published article and its supplementary information files.
-
The authors declare that they have no conflict of interest.
- Copyright: © 2024 by the author(s). Published by Maximum Academic Press on behalf of China Agricultural University, Zhejiang University and Shenyang Agricultural University. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Zhang Z, Shen Y, Xin G, Deng W, Tan H, et al. 2024. The effect of static magnetic field on inducing the binding of bovine serum albumin and cyanidin-3-O-glucoside. Food Innovation and Advances 3(4): 449−456 doi: 10.48130/fia-0024-0042
The effect of static magnetic field on inducing the binding of bovine serum albumin and cyanidin-3-O-glucoside
- Received: 28 June 2024
- Revised: 11 December 2024
- Accepted: 11 December 2024
- Published online: 24 December 2024
Abstract: Serum albumin can bind with a diverse range of small molecules. It could therefore serve a protective or carrier function, and effectively address the issue of anthocyanins' susceptibility to decomposition. The anisotropic effect of the magnetic field (MF) can influence their interaction, thereby playing a distinct role in molecular bonding. In this study, bovine serum albumin (BSA) and cyanidin-3-O-glucoside (C3G) were used as raw materials. The mechanism underlying the formation of BSA-C3G complexes induced by static magnetic field (SMF) was investigated through analyses of secondary structure, functional groups, dipole moment, crystal cell dimensions, and microstructural characteristics. BSA and C3G were treated with 50, 100, 150, and 200 mT, respectively. As the magnetic intensity increased, the secondary structure of the complex changed, the α-spiral content, β-corner content, and irregular curl content decreased, while, the β-folding content increased. The average grain size of the BSA-C3G composite was observed to decrease. Furthermore, alterations in the crystal cell dimensions of the BSA-C3G complex were noted, accompanied by a tendency for the microstructure to become more flattened. This study offers valuable insights into the influence of SMF on the assembly behavior and structural characteristics of proteins and anthocyanins.
-
Key words:
- Static magnetic field /
- Anthocyanins /
- Bovine serum albumin /
- Complexes /
- Molecular binding