-
Mirabilis himalaica (Edgew.) Heimerl, commonly known as Ba Zhu in Xizang, belongs to the Nyctaginaceae family. It is the only Old World representative of a large New World genus[1]. The distribution of M. himalaica is confined to the dry-hot valleys of the Qinghai-Xizang Plateau, although it can thrive in areas with significant altitude variations, demonstrating remarkable altitude adaptability[2]. Native Xizang people have used M. himalaica since the Tang Dynasty to treat stomach disorders, nephritic edema, and gonorrhea[3]. Its limited distribution, combined with the growing demand for traditional medicinal plants, puts pressure on wild populations of M. himalaica. According to the IUCN Red List Categories and Criteria (version 3.1)[4] and the Guidelines for Application of IUCN Red List Criteria at Regional Levels (version 3.0)[5], M. himalaica is classified as 'Near Threatened' in China, based on field surveys[1]. The ecological niche and medicinal value of M. himalaica have garnered attention, prompting conservation efforts and research on its bioactive compounds. Artificial cultivation of M. himalaica was conducted in Xizang, although cultivated and wild populations exhibit differences in chemical composition[6]. Numerous active compounds have been identified in M. himalaica, most of which are phenolic acids, particularly rotenoid-type flavonoids[7,8]. For example, Linghu et al. isolated Mirabijalone E, which exhibited anticancer properties[8]. Other notable properties, such as myxospermy[9] and UV-B radiation adaptation[10], have also been investigated.
Genetic transformation is a critical tool for investigating the biosynthesis pathways of species-specific bioactive compounds, unique structural adaptations for plateau conditions, and genetic improvement. However, genetic transformation is limited for M. himalaica. Lan et al. established hairy-root transformation of M. himalaica[3]. Currently, there have been no reports on methods for obtaining transgenic regenerated M. himalaica through genetic transformation. Agrobacterium tumefaciens-mediated transformation is the most widely used method for genetic transformation; however, it is often inefficient, requiring the cumbersome process of tissue culture and is species- and genotype-dependent[11]. Thus, developing non-tissue culture transformation and regeneration systems is critical in plant science research, especially for non-model plants. Agrobacterium rhizogenes-mediated non-tissue culture transformation is an attractive strategy[12]. A. rhizogenes-mediated transformation has higher efficiency and better plant species adaptability compared with A. tumefaciens-mediated transformation[13]. The limitation of A. rhizogenes-mediated transformation is that it is generally used to generate transgenic hairy roots, which are difficult to convert into shoots. Recently, Cao et al. established a simple cut-dip-budding (CDB) delivery system, which enabled efficient transformation under non-sterile conditions without the need for tissue culture[11]. This method has subsequently been applied to Chinese cabbage[14], Idesia polycarpa[15], succulent plants[16], trees[17], as well as medicinal plants[18].
Due to the special ecological niche and medicinal value of M. himalaica, we established a simple and efficient A. rhizogenes-mediated transformation system. Using leaves as explants, we successfully transformed a fluorescence and betacyanin reporter system into M. himalaica, obtaining transgenic plants. This is the first report of the transformation of a Xizang folk medicinal plant that thrives at high altitudes (over 3,000 m). Our work expands the application of the cut-dip-budding (CDB) system for plant transformation and will promote investigations into the biosynthetic pathways of bioactive compounds and genetic improvement in Xizang medicinal plants.
-
The seeds of M. himalaica were collected in Linzhi, Xizang Autonomous Region, China. These seeds were germinated in Petri dishes on wet filter paper for 2 d and then grown in an artificial climate chamber (24 °C, 16 h light/22 °C, 8 h dark). Leaves from 1-month-old M. himalaica were used for transformation.
Preparation of Agrobacterium rhizogenes
-
The reporter constructs, pYL1300H-CDGAeG[19] and Cotton 2.0-tdTomato[20], used in this study were kindly provided by Prof. Qinglong Zhu (South China Agricultural University, Guangzhou, China) and Prof. Lu Long (Henan University, Kaifeng, China), respectively. In pYL1300H-CDGAeG, four betacyanin biosynthetic genes, including BvADH, BvCYP76AD1S, BvDODA1S, and cDOPA5GT, as well as an eGFP gene, are expressed under the control of 35S promoters. In Cotton 2.0-tdTomato, a red fluorescence gene, tdTomato, which is significantly brighter than eGFP, is expressed under the control of the enhanced 35S promoter.
The plasmids were introduced into A. rhizogenes K599 cells via heat-shock transformation, as described by Cao et al.[11]. The transformed Agrobacteria were cultured (28 °C, 220 rpm) to reach an OD600 of 0.8–1 in TY medium (5 g/L tryptone and 3 g/L yeast extract) containing 50 mg/L streptomycin, 50 mg/L kanamycin, and 10 mM CaCl2. Three hundred microlitres of this culture were spread and cultured for 2~3 d on TY solid agar medium (5 g/L tryptone, 3 g/L yeast extract, and 15 g/L agar) until the agar medium was covered with a uniform layer of bacteria. The bacterial suspension in liquid TY medium and the slush bacterial layer from the solid agar medium were then prepared for plant infection.
Plant transformation
-
In this study, we modified the cut-dip-budding (CDB) method reported by Cao et al.[11]. Briefly, mature leaves from 1-month-old M. himalaica were cut from the petiole and used as explants. The cut petiole ends were vacuum-infiltrated for 5 min with an A. rhizogenes K599 suspension prepared in an infection solution containing 10 mM MgCl2, 10 mM MES, and 100 μM acetosyringone (pH 6.0). Subsequently, the cut ends of the petioles were coated with bacterial layers from the TY solid agar medium. These inoculated leaves were then planted in pots containing vermiculite and covered with plastic wrap to maintain moisture. The inoculated explants were cultured under plastic wrap at 26 °C with a 16-h light/8-h dark cycle until buds developed from the leaf cuttings (approximately 1 month).
Verification of the transgenic M. himalaica
-
The transgenic buds were first examined for purple color (indicative of the betacyanin reporter system) or red fluorescence (indicative of the tdTomato reporter system) under the 538 nm wavelength (LUYOR-3410GR). To further verify the presence of transgenic plants, genomic DNA was isolated from young leaves of M. himalaica using the CTAB method and used as templates for PCR. The primers used in genomic PCR are listed in Supplementary Table S1.
Expression analysis of exogenous genes
-
To detect the expression levels of exogenous genes, we extracted total RNA from the wild-type and transformed regenerated M. himalaica, and reverse transcribed it into cDNA. Then it was used as a template for q-RTPCR. Beta-actin was used as an housekeeping gene in M. himalaica. The qRT-PCR primers are shown in Supplementary Table S1.
Determination of betanin content
-
To extract betanin from M. himalaica, 1.0 g of fresh leaf tissue was ground into a fine powder using liquid nitrogen. The powdered tissue was then transferred to an Eppendorf tube containing 10 mL of distilled water. The tube was placed in an ultrasonic cleaner and maintained at 40 °C for 60 min. Afterward, the mixture was centrifuged at 8,000 rpm for 10 min, and the supernatant was collected for the comparison of betanin contents.
To detect betanin, a Shimadzu LC-20A HPLC system with a photo-diode array detector was used (Shimadzu, Kyoto, Japan). The separation was carried out on a Kromasil-C18 column (4.6 mm × 250 mm, i.d., 5 μm) with methanol (A)-formic acid aqueous solution (0.2%, B) as the mobile phase. The isocratic elution was performed at a flow rate of 0.8 ml/min for 10 min (A:B = 15:85 v/v). The detection wavelength was 535 nm, the column temperature was 40 °C, and the injection volume was 10 μL. The authentic betanin was purchased from Shanghai yuanye (Shanghai yuanye Bio-Technology Co., Ltd, Shanghai, China). The standard curve and linear regression equations are provided in Supplementary Fig. S1.
-
The selection of adaptive explants is crucial for non-tissue culture transformation. To identify the most suitable explants for transformation, we focused on the regeneration capabilities of different tissue parts of Mirabilis himalaica. We conducted experiments using three types of explants: root segments, stem segments, and leaves. The results indicated that after approximately 10 d, the stem segments successfully developed adventitious roots; however, they did not produce any regeneration buds even after 40 d. Root segments failed to produce either buds or roots. In contrast, the leaves of M. himalaica demonstrated robust regenerative potential, rooting and surviving within about 10 d. After approximately 30 d, adventitious buds began to emerge at the base of the petioles, with each petiole averaging 1.3 adventitious buds (Table 1). Furthermore, experiments investigating petioles of varying ages—mature and juvenile—showed that while both types of leaves could root, the mature petioles exhibited superior performance in both rooting and budding.
Table 1. Statistics on leaf regeneration and buds emergence of Mirabilis himalaica.
Experiment No. of explants No. of buds Ratio of buds/explants I 114 148 1.30 II 135 177 1.31 III 100 132 1.32 In comparison to other tissue parts used as explants, petiole leaves offer accessibility and convenience, which is particularly significant for large-scale genetic transformation, ensuring a reliable source of explants. In conclusion, the mature leaves of M. himalaica exhibited robust regeneration and budding capacity, making them ideal explants for the non-tissue culture transformation of M. himalaica.
Establishment of transformation system using leaf petioles as explants
-
To establish a non-tissue culture transformation system using leaf petioles as explants (Fig. 1a), we employed A. rhizogenes K599 carrying the pYLTAC380H-CDGAeG construct, in which four betacyanin biosynthetic genes and an eGFP gene were assembled using the TGSII-UNiE system[19]. The petioles of M. himalaica were immersed in the infection solution and placed in a vacuum chamber for 5 min (Fig. 1b). Subsequently, we inoculated the leaf petioles with A. rhizogenes K599 (Fig. 1c) and inserted them into moist vermiculite for cultivation (Fig. 1d & e). Hairy roots were induced after approximately 10 d, and budding emerged after 30 d (Fig. 1f & g).
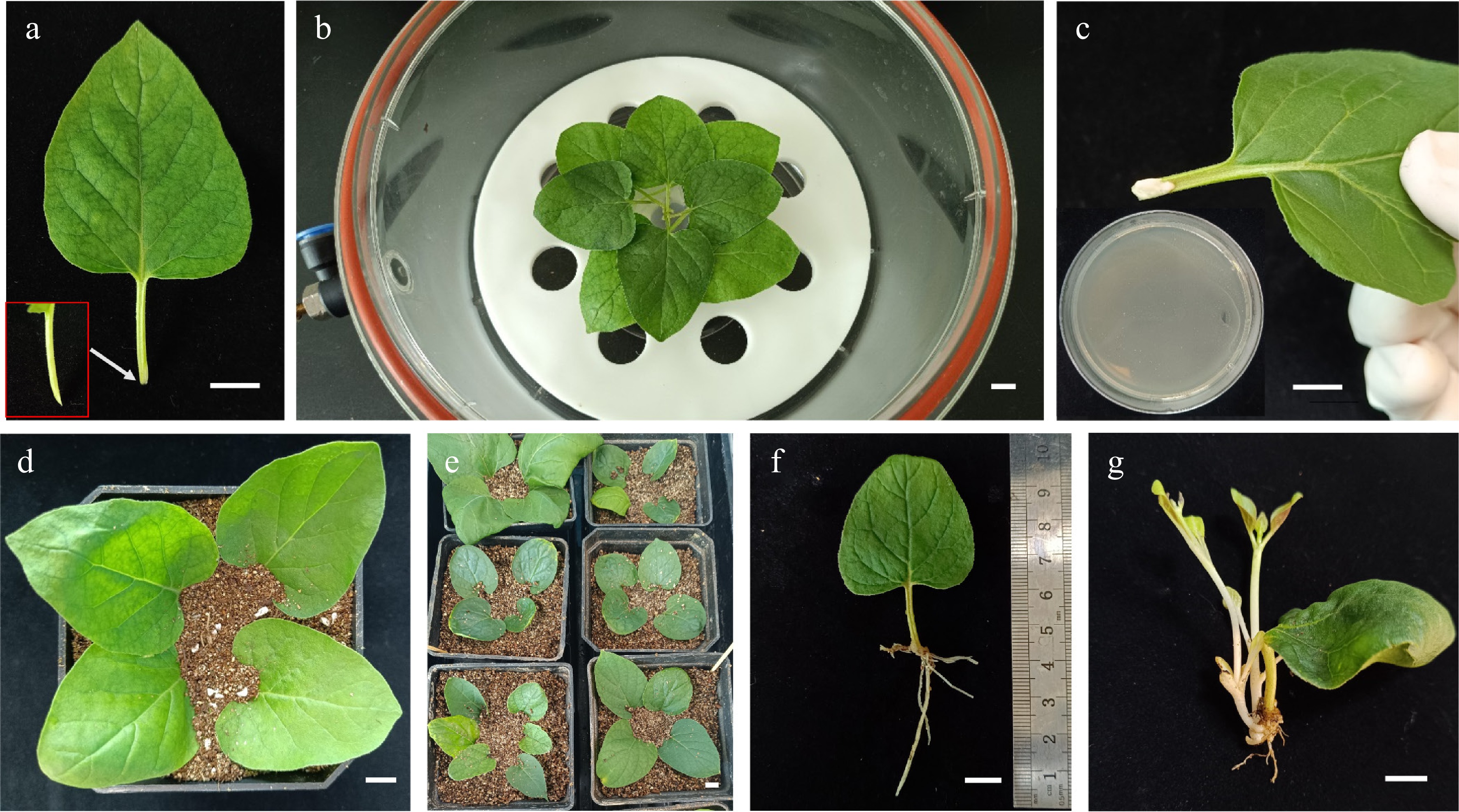
Figure 1.
Genetic transformation of Mirabilis himalaica using the modified CDB protocol. (a) Petiolate leaves were cut and used as explants. The site of infection by A. rhizogene was enlarged. (b) The explants soaked in A. rhizogenes K599 suspension were then subjected to vacuum pressure. (c) Coating the cut sites with A. rhizogenes K599 layers from agar media. (d)−(e) Explants inoculated with A. rhizogenes K599 were cultured in soil. (f) The root of 10 d old explants. (g) Buds emerged after about 30 d. Scale bars: 1 cm.
The red color of betacyanin was used to monitor transformation events. Compared to the wild type, the buds and roots of positive transformants of M. himalaica accumulated higher levels of betacyanin (Fig. 2a−d). To confirm the transformation of M. himalaica, we conducted genomic PCR to examine the presence of exogenous, rolB and rolC genes. As shown in Fig. 2e, four betanin biosynthetic genes, rolB and rolC gens were amplified in the transgenic M. himalaica plants. This indicates that the A. rhizogenes-mediated transgenic process has successfully inserted the target genes into M. himalaica genome. The leaves of transgenic M. himalaica plants are redder than those of the wild type (Fig. 2f). Simple extraction of betacyanin using water also indicates that the genetically modified M. himalaica may accumulate more betacyanin (Fig. 2g). To further confirm whether the genetically modified M. himalaica accumulates more betanin, we used qRT-PCR to detect the expression levels of genes involved in betanin biosynthesis and employed HPLC to measure the content of betanin. The expression levels of four betanin biosynthetic genes were significantly increased in the transgenic plants (Fig. 2h−k). The betanin contents in transgenic M. himalaica is 5−6 folds higher compared with that in the wild type (Fig. 2l & Supplementary Fig. S2). To analyze the genetic transformation efficiency using this procedure, we repeated the genetic transformation experiments three times, using 100 explants each time. The average rooting rate was about 89%, and the rate of positive roots (exhibiting red color) was also about 78%. However, the rate of positive buds was relatively low, at only 1.7% of the total explants (Table 2).
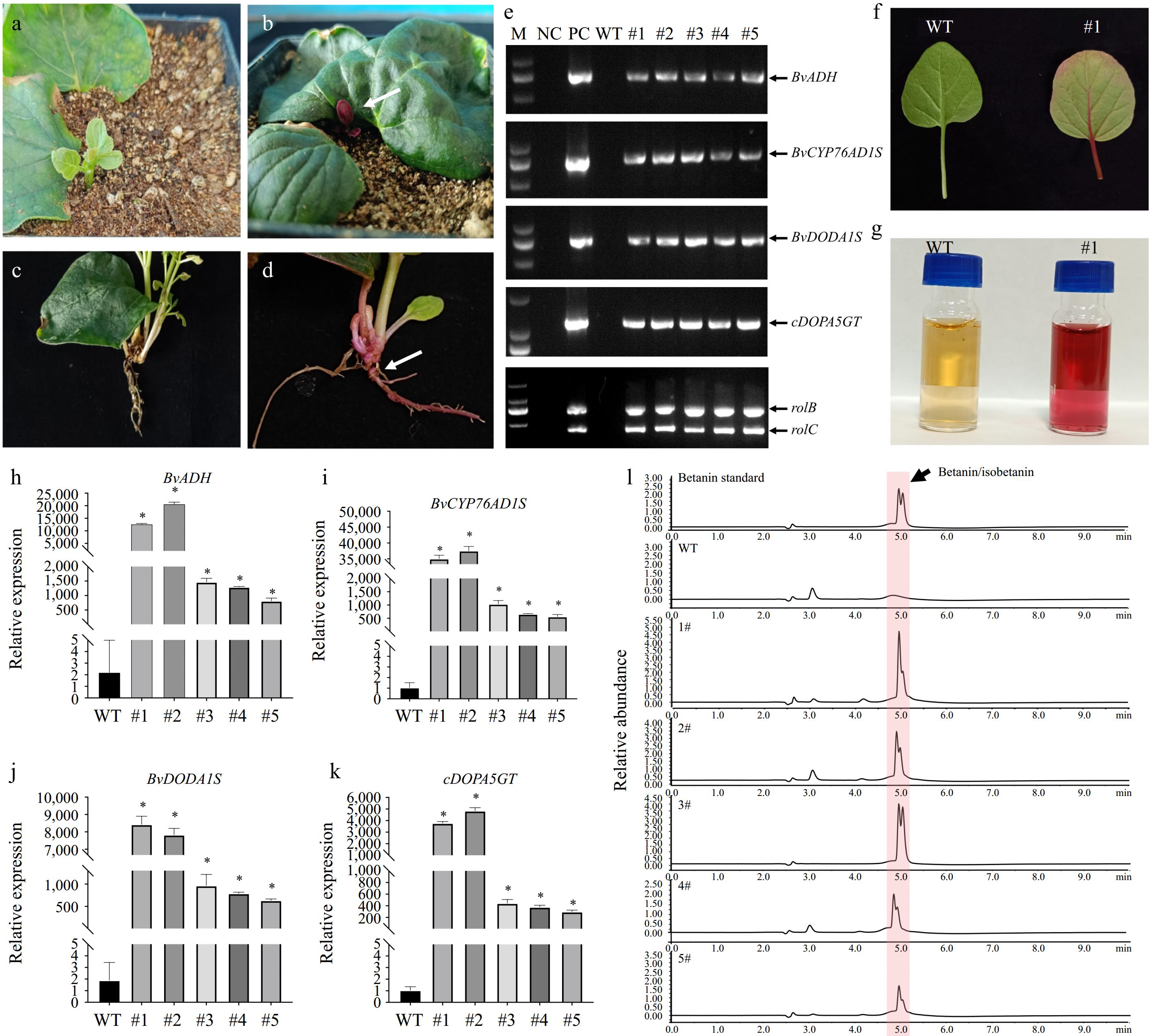
Figure 2.
Genetic transformation of M. himalaica using betacyanin as a reporter. (a) & (b) The above-ground part of M. himalaica, showing the (a) non-transgenic, and (b) transgenic. White arrow indicates red bud with high betacyanin contents. (c) & (d) Roots of M. himalaica. White arrow indicates red roots with high betacyanin contents. (e) Genomic PCR detect four exogenous genes. M: marker; NC: negative control (sterilized water was used as templates); PC: positive control (plasmid pYL1300H-CDGAeG was used as templates); WT: wild type M. himalaica. (f) The betacyanin accumulated in pYL1300H-CDGAeG transgenic M. himalaica leaf. (g) The leaf of pYL1300H-CDGAeG transgenic M. himalaica (right) have higher betacyanin contents compared with wild type M. himalaica (left). (h)−(k) Relative expression levels of betanin biosynthetic genes by qRT-PCR in transgenic M. himalaica, respectively, compared with wild-type (WT) M. himalaica. All data represent the mean ± SD (n = 3; *, p < 0.001; Student's t-test). (l) Analysis of betanin in leaves of M. himalaica by HPLC.
Table 2. Statistics of M. himalaica transformation efficiency using betacyanin as a reporter.
Experiment Explants Rooting explants Transgenic roots Transgenic plants Rooting rate Positive rooting rate Positive plant rate I 100 84 75 2 84% 75% 2% II 100 90 80 1 90% 80% 1% III 100 92 79 2 92% 79% 2% Establishment of transformation system using tdTomato as a reporter
-
Using betacyanin as a reporter requires at least three genes, which complicates plasmid construction and may also reduce transformation efficiency. In addition, many plants in Caryophyllales, such as M. himalaica, also produce betacyanin which might interfere the judgement. Therefore, we employed tdTomato as a reporter, an exceptionally bright red fluorescent protein that is about six times brighter than eGFP. We used A. rhizogenes K599 carrying a construct in which tdTomato was driven by the 35S promoter to infect Mirabilis himalaica leaves following the same protocol. The transgenic plants were checked using a portable fluorescent lamp (Fig. 3a−d). The transformants were further confirmed by genomic PCR (Fig. 3e). Further qRT-PCR results showed that the expression levels of tdTomato gene was increased in the transgenic plants (Supplementary Fig. S3). We conducted three repeated experiments as well, using 100 explants each time. The positive rooting rate and the positive plant rate were similar to those obtained using betacyanin biosynthetic genes, at 73% and 2%, respectively (Fig. 3f).

Figure 3.
Genetic transformation of M. himalaica using tdTomato as a reporter. (a) & (b) Screening positive plants with a portable fluorescent lamp. Two representative plants growing closely together are displayed here. The plants enclosed in solid rectangular boxes are positive plants and arrow with solid line showed the fluorescent elicited by tdTomato protein. The plants enclosed in dotted rectangular boxes were negative transgenic M. himalaica and the arrow with dotted line indicated negative fluorescent signal. (c) & (d) The fluorescent of positive tdTomato transgenic plants. The roots enclosed in dotted rectangular boxes represented negative transgenic roots and the arrows with solid line showed the fluorescent elicited by tdTomato protein. (e) Genomic PCR detect the tdTomato, rolB and rolC genes in transgenic plants. M: marker; NC: negative control (sterilized water was used as templates); PC: positive control (plasmid Cotton 2.0-tdTomato was used as templates); WT: wild type M. himalaica. (f) Statistics of M. himalaica transformation efficiency using tdTomato as a reporter. Scale bar: 1 cm.
-
In this study, we developed a simple non-tissue culture transformation system for the Xizang folk medicinal plant Mirabilis himalaica using Agrobacterium rhizogenes-mediated genetic transformation technology. This method has been successfully applied to medicinal plants, as it overcomes many challenges associated with the Agrobacterium tumefaciens-mediated transformation method, including long transformation periods, rigorous sterile conditions, and limitations regarding species and genotype[15,18]. Due to the unique growth habits of plateau plants, a stable genetic transformation system has not yet been established for many Xizang medicinal plants[3]. In our study, leaves were first selected as explants. Subsequently, we successfully obtained transgenic Mirabilis himalaica via A. rhizogenes-mediated genetic transformation. This procedure is simple and does not require tissue culture. Furthermore, using leaves as explants is advantageous because they are the most abundant tissues in plants and are relatively easy to obtain. The establishment of this transformation procedure for Mirabilis himalaica lays a solid foundation for future gene function identification through gene overexpressing or editing[21,22]. Furthermore, the roots of M. himalaica can also serve as a chassis for synthetic biology, similar to the endosperm of rice[23].
The reporter is crucial for monitoring genetic transformation, especially in non-tissue culture procedures. Unlike A. tumefaciens mediated tissue culture transformation, antibiotics are not the preferred selection markers for non-tissue culture transformation[12]. Pigments, such as betacyanin, and fluorescent proteins are the most commonly used reporters for the visualization of transformation[15,24]. However, plants belonging to the Caryophyllales order produce betacyanins pigments themselves. The endogenous betacyanin may interfere with the identification of transformants when betacyanin is used as a reporter. Thus, betacyanin is not a preferred reporter for Caryophyllales plants. In this study, we used tdTomato as an alternative reporter, which is smaller than the four betacyanin biosynthetic genes or the RUBY reporter. This simplification aids in plasmid construction and is suitable for large-scale genetic transformation. Nevertheless, the fluorescent signal was relatively weak in the leaves of transgenic M. himalaica in our study (Fig. 3b). The presence of pigments, such as chlorophyll and betacyanin, might hinder the fluorescent signal. Notably, betaxanthins, the intermediates of betacyanin, emit visible green fluorescence under blue light excitation[25]. More suitable fluorescent proteins should be screened to improve the visualization of transformations in Caryophyllales plants. An alternative reporter could be herbicide resistance genes, such as the BAR or EPSPS gene. For example, phosphinothricin (Basta) has been used for bulk selection of transgenic sweet potato[24].
Our study demonstrated that the transgenic adventitious roots of Mirabilis himalaica could produce shoots. The method we used is similar to the recently reported cut-dip-budding (CDB) method[11]. Using this approach, transformants have been successfully obtained from several other plants, including two herbaceous plants (Taraxacum kok-saghyz and Coronilla varia), a tuberous root plant (sweet potato), and three woody plant species (Ailanthus altissima, Aralia elata, and Clerodendrum chinense)[11]. More recently, transformation systems for some medicinal plants, including Pugongying (Taraxacum mongolicum), Dihuang (Rehmannia glutinosa), Danshen (Salvia miltiorrhiza), and Yuanzhi (Polygala tenuifolia), have been established using this method[18]. Compared to the results reported by Cao et al.[11], the rate of positive plants in our study was relatively low (Fig. 3f, Table 2), averaging 2%. In addition to potential species differences, several factors should be considered in future studies to increase the conversion rate from roots to shoots, such as environmental conditions and plant hormones. Nonetheless, the establishment of a genetic transformation system for Mirabilis himalaica is beneficial and will facilitate the development of genetic transformation systems for other Xizang medicinal plants.
This work was financially supported by the National Natural Science Foundation of China (Grant No. U20A20401). Thanks to Prof. Qinglong Zhu (South China Agricultural University, Guangzhou, China) and Prof. Lu Long (Henan University, Kaifeng, China) who kindly provided pYL1300H-CDGAeG and Cotton 2.0-tdTomato constructs.
-
The authors confirm contribution to the paper as follows: study conception and design: Zhang F, Lan X; data collection: Sun T, Han X, Jiang Y, Li Q, Xu Y; analysis and interpretation of results: Zhang F, Han X; draft manuscript preparation: Lan X, Sun T. All authors reviewed the results and approved the final version of the manuscript.
-
All the data generated or analyzed during this study are included in this published article and its supplementary information files.
-
The authors declare that they have no conflict of interest. Dr. Fangyuan Zhang is the Editorial Board member of Medicinal Plant Biology who was blinded from reviewing or making decisions on the manuscript. The article was subject to the journal's standard procedures, with peer-review handled independently of this Editorial Board member and the research groups.
- Supplementary Table S1 Primers used in this study.
- Supplementary Fig. S1 The standard curve between the HPLC peak area and betanin content (0, 1.5, 2, 3, 4, and 5 mg/ml). A linear regression was observed between the peak area and betanin contents at 0−5 mg (R2 = 0.9865).
- Supplementary Fig. S2 The contents of betanin in pYL1300H-CDGAeG transgenic plants and wild type M. himalaica. All data represent the mean ± SD (n = 3; *, p < 0.001; Student's t-test).
- Supplementary Fig. S3 Relative expression levels of tdTomato gene by qRT-PCR in tdTomato overexpression plants, respectively, compared with that in wild type (WT) M. himalaica. β-actin was used as an internal reference gene. All data represent the mean ± SD (n = 3; *, P < 0.001; Student's t-test).
- Copyright: © 2025 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Sun T, Han X, Jiang Y, Li Q, Xu Y, et al. 2025. A method of genetic transformation of Xizang medical plants without tissue culture. Medicinal Plant Biology 4: e002 doi: 10.48130/mpb-0024-0032
A method of genetic transformation of Xizang medical plants without tissue culture
- Received: 05 November 2024
- Revised: 07 December 2024
- Accepted: 13 December 2024
- Published online: 20 January 2025
Abstract: Mirabilis himalaica, known for its medicinal properties and limited distribution in the Qinghai-Xizang Plateau, is under conservation pressure due to high market demand. Traditional transformation methods are inefficient for this species, prompting the development of a non-tissue culture approach. This study utilized mature leaves as explants and employed a cut-dip-budding (CDB) system to transform fluorescence and betacyanin reporter systems in M. himalaica, successfully transforming a high-altitude Xizang medicinal plant. The study found that mature leaves exhibited robust regeneration potential, making them ideal for large-scale transformation. The transformation system achieved approximately 76% positive rooting rate and a 2% positive transgenic plant rate. This method overcomes challenges associated with Agrobacterium tumefaciens-mediated transformation and lays the foundation for future genetic research and industrial applications of Xizang medicinal plants.













