-
Angiosperms are the most successful group of land plants that have colonized virtually all terrestrials on this planet[1]. This evolutionary success largely depends on two related morphological innovations, seed production via sexual reproduction and the origin of fruits that facilitate seed dispersal[2−6]. A successful sexual reproduction process requires both the development of male and female gametes through meiosis, during which genetic variations are generated that are potentially adaptive in a new environment[7]. Meanwhile, the fruits provide an enclosed, stable condition for the developing seeds, and when the seeds mature, the fruits encourage the seeds to excavate new habitat through distinct dispersal strategies[4,8−11]. The fruit is derived from the gynoecium consisting of one or more carpels, which is the female reproductive organ that develops in the center of a flower[2,12,13]. In addition to its evolutionary contribution to angiosperm diversification, the fruits are also the source of vitamins, proteins, fibers, and carbohydrates for human diet. Deciphering the genes and genetic networks contributing to the gynoecium patterning and fruit development has been a central topic in plant biology, not only for a better understanding of the mechanism underlying plant diversification but also for providing the blueprint for crop improvement.
The gynoecium is inarguably the most complex plant organ consisting of multiple tissues and distinct cell types elaborated along the three axes: (1) the differentiation of gynophore, ovary, style, and stigma along the proximal-distal axis; (2) the replum, placenta tissues, valve margin, and valves along the medial-lateral axis; and (3) the differentiation of endocarp, mesocarp, and exocarp layer in the valves along the adaxial-abaxial axis, respectively (Figs 1, 2)[12−15]. The correct patterning of these cells/tissues requires the precise territorial expression of key transcription factors in the gynoecium morphogenesis process[2,14−18]. In addition, recent findings also suggest the involvement of plant hormones in regulating gene activities and expression during gynoecium and fruit development[19,20]. Altogether, these factors constitute robust genetic regulatory networks (GRNs) to ensure the correct patterning of the distinct tissues in the gynoecium and the successful development of a fruit[2,18,19].
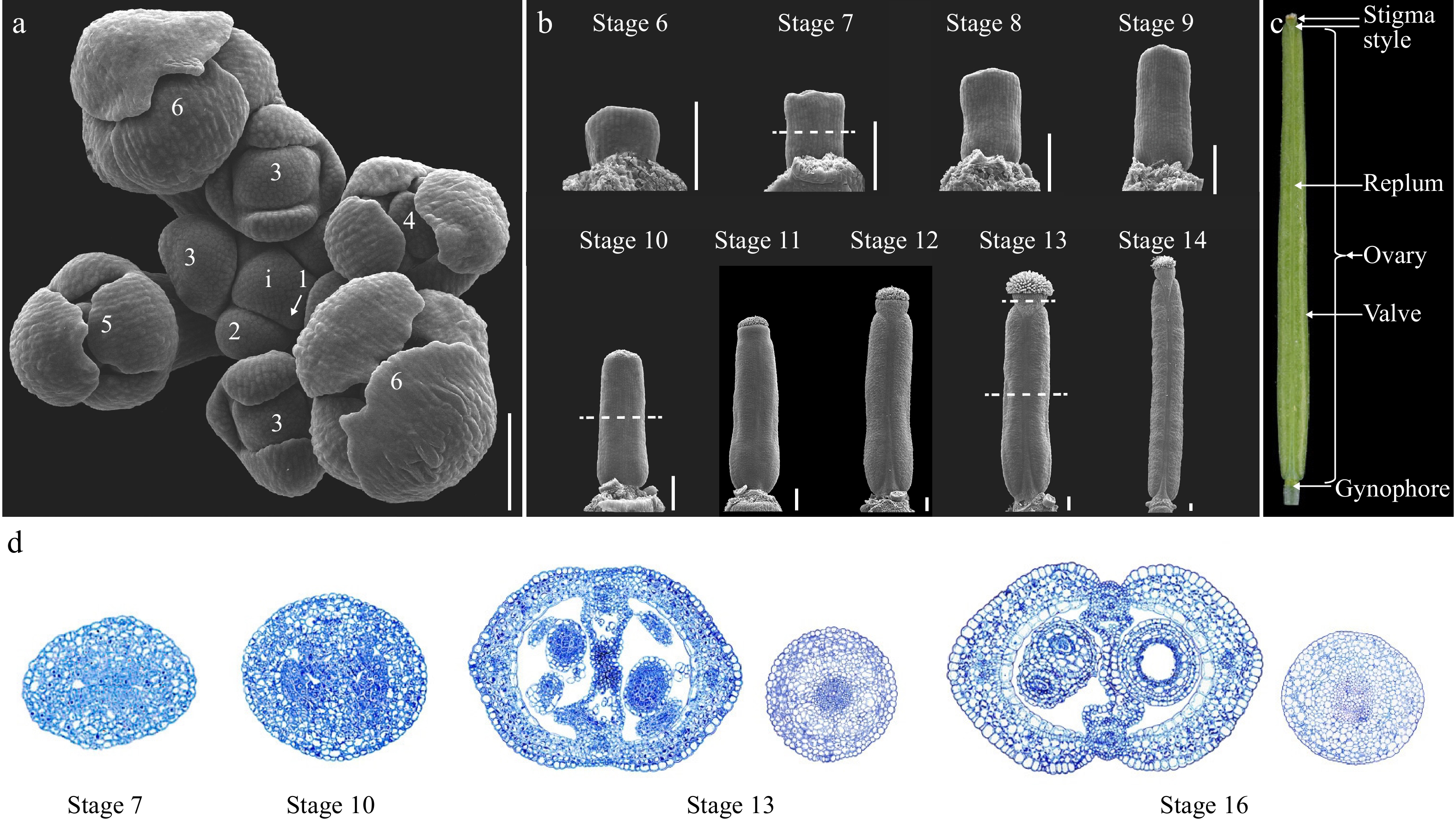
Figure 1.
Overview of the gynoecium development in Arabidopsis. (a) Top view of an Arabidopsis inflorescence meristem (IM) consisting of stage 1 to stage 6 floral primordia. Dashed lines indicate the position of the cross section shown in (d). (b) SEM micrographs of Arabidopsis gynoecium from stages 6 to 14. (c) A mature Arabidopsis fruit at stage 17 showing the full differentiation of distinct tissues. (d) Cross-section of the gynoecium at stage 7, 10, 13, and 16. For stage 13 and 16 samples, both the ovary and style were included for comparison of the symmetry. i: inflorescence meristem; the numbers indicate the developmental stages. Scale bars = 100 μm.
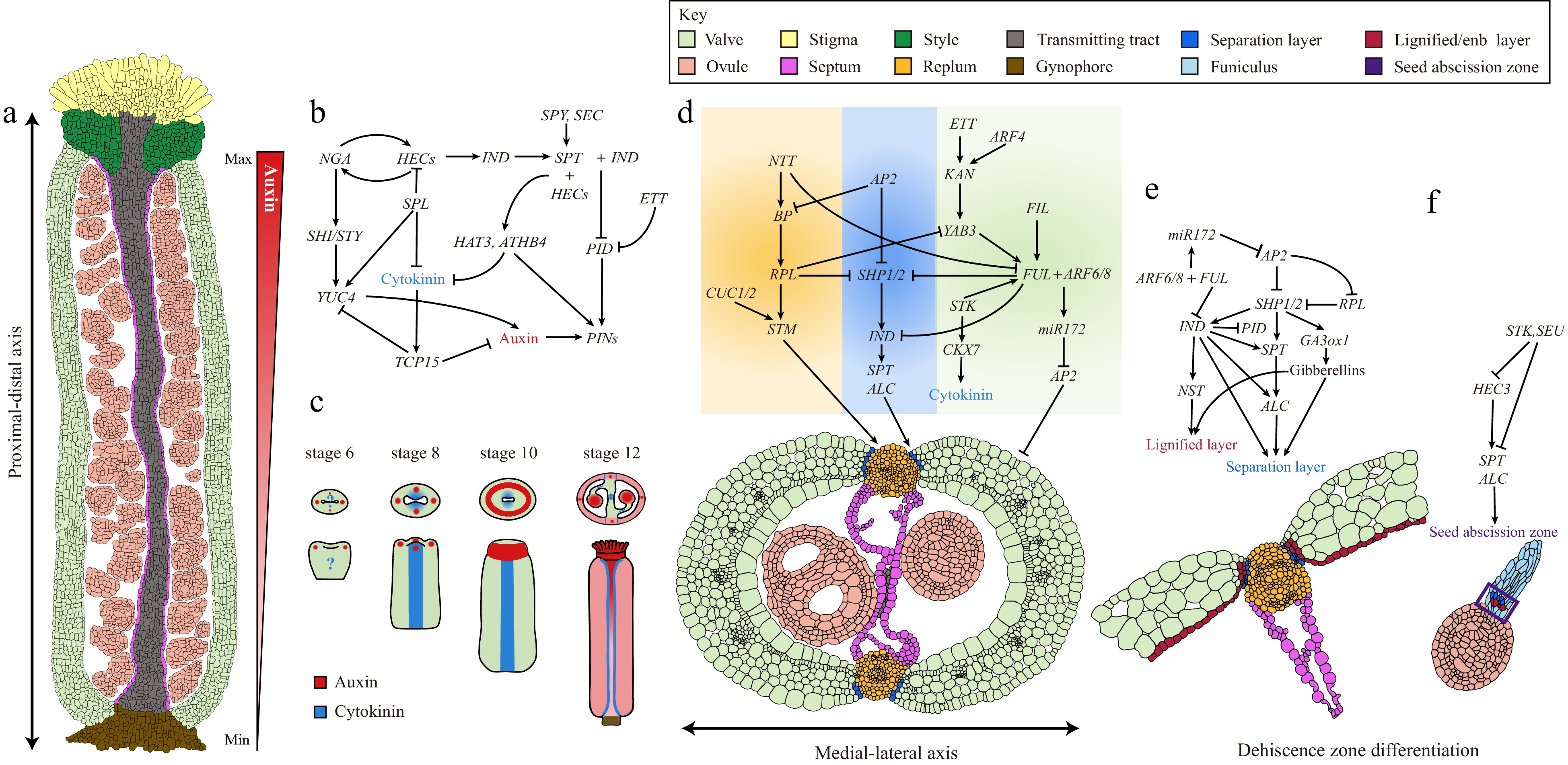
Figure 2.
Complex GRNs underlie gynoecium patterning and fruit development in Arabidopsis. (a) A longitudinal section of a stage 11/12 Arabidopsis gynoecium showing the differentiation of complex tissues along the proximal-distal axis. The auxin gradient along the proximal-distal axis is shown by a triangular red gradient. (b) GRNs and the hormone dynamic patterning the gynoecium apical region. (c) Auxin and cytokinin signaling distribution pattern during gynoecium development from stage 6 to 12. (d) GRNs governing the differentiation of medial-lateral axis of Arabidopsis gynoecium. (e) GRNs and the hormone dynamic controlling the dehiscence zone differentiation. (f) GRNs controlling the cell differentiation responsible for seed abscission.
In this review, the key landmarks in gynoecium patterning and fruit development in Arabidopsis were integrated into a developmental framework, within which we discuss the regulatory networks underlying these key developmental steps. The role of plant hormones are also summarized, especially auxin and cytokinin, in gynoecium patterning and fruit development, respectively. The key components in gynoecium/fruit development that can be potentially utilized for crop improvement by modern genetic toolkits are also highlighted.
-
Arabidopsis is a representative of Brassicaceae, whose gynoecium is composed of two congenitally fused carpels in the middle of the flower[12,13]. The mature fruit is a dehiscent dry fruit specialized to Brassicaceae, termed as silique. The whole developmental process of the Arabidopsis flower can be meticulously divided into 20 sequential stages with stages 6 to 13 related to gynoecium patterning whilst stages 14 to 20 encompasses the fruit morphogenesis process after pollination (Fig. 1a−c)[13]. The gynoecium primordium is initiated as an elliptical flattened protrusion in the middle of the floral meristem at stage 6. From stages 6 to 8, the gynoecium primordium continues to grow both longitudinally and laterally as a hollow tube, and at stage 9, the style and stigmatic papilla cells start to differentiate (Fig. 1b, d)[13]. The gynoecium continues to grow longitudinally from stages 10 to 12. At late stage 12, the valves, valve margins, replum, and style start to become morphologically evident, and the gynoecium is ready for fertilization at stage 13 (Fig. 1b, d)[13]. The fruit developmental program starts from stage 14 upon the fertilization of the egg cells. The fruit elongates dramatically from stage 14 to 16 and reaches the final size at stage 17 (Fig. 1c)[13]. At later stage 17, the valve margin begins to differentiate into the dehiscence zone, and lignified cells in the valve margin and endocarp b (enb) layer starts to deposit secondary cell walls (Figs 1c, 2)[13]. Meanwhile, the middle lamella of the separation layer starts to break down. All these developmental and physiological processes make the silique ready for dehiscence. The pods turn yellow and finalize the shattering process from stages 18 to 20[13].
-
Plants continuously generate lateral organs to sustain their growth and development throughout their lifecycle[21]. This feature is largely attributable to the activity of the shoot apical meristem (SAM) located at the very tip of a shoot[22]. In the SAM, a group of stem cells sustains the pluripotent activity of the meristem so that new organs are repeatedly produced[23]. Under certain conditions, such as long day light periods and vernalization, the SAM transits into the inflorescence meristem (IM), on which flanks the floral meristems (FMs) are produced[24]. During the development of an FM, four types of floral organs are produced in whorls: sepals, petals, androecium, and gynoecium. These floral organs are specified by combinatorial protein complexes of floral organ identity genes in a framework known as the ABC model[25,26]. The gynoecium identity itself is defined by the quaternary protein complex constituting two AGAMOUS (AG) with two SEPALLATA (SEP) proteins. Single ag loss-of-function mutation or multiple sep gene mutation results in the loss of gynoecium identity[27,28].
Unlike the SAM and IM, which are indeterminate, the FM is determinate by the initiation of two carpel primordia. In both SAM and IM, the indeterminacy is achieved by the maintenance of stem cells, which is regulated by the negative feedback loops between the homeodomain transcription factor WUSCHEL (WUS) and the CLAVATA 3 (CLV3) ligand-receptor system[29,30]. In the SAM, WUS positively regulates stem cell maintenance and activates the expression of CLV3, which in turn restricts the expression domain of WUS[31,32]. The establishment of gynoecium identity by AG is required for the FM termination as ag mutation changes the determinate gynoecium primordium into an indeterminate meristem with repeated floral organ initiation[27]. At the very early developmental stages of an FM, both WUS and CLV3 can be detected in the center of the FM, where WUS together with LEAFY (LFY) activates the expression of AG at stage 3 and onward[33]. Indeed, the expression of AG and the gynoecium identity is diminished in either weak wus mutant or null lfy mutant, respectively[33]. In addition, the bZIP transcription factor PERIANTHIA (PAN) was also identified as a positive regulator of AG[34]. AG is down-regulated in the fourth whorl of the pan mutant and results in an enlarged meristem consequent upon the extension of WUS expression (Fig. 3; Table 1)[34,35].

Figure 3.
Expression patterns of the genetic factors involved in gynoecium patterning. A visual depiction of the expression patterns of genetic factors involving in the development and specification of gynoecium tissues. Longitudinal and cross sections of the gynoecium at stages 6, 10, and 13 are shown. Expression is indicated by the red color exclusively within the gynoecium tissues (gene expression patterns are derived from original research articles; the relevant sources can be found in Table 1).
Table 1. A list of genes involved in the gynoecium patterning and fruit development.
Gene name AG code Family Function Ref. AGAMOUS (AG) AT4G18960 MADS-box Gynoecium primordium specification [27] ALCATRAZ (ALC) AT5G67110 bHLH Separation layer development [117] APETALA 2 (AP2) AT4G36920 AP2/EREBP Valve development [110,120] ARABIDOPSIS RESPONSE REGULATORS 1(ARR1) AT3G16857 ARR CMM development [83] ARABIDOPSIS RESPONSE REGULATORS 10 (ARR10) AT4G31920 ARR CMM development [83] ARABIDOPSIS RESPONSE REGULATORS 12 (ARR12) AT2G25180 ARR CMM development [83] AUXIN RESPONSE FACTOR 3/ ETTIN (ARF3/ETT) AT2G33860 ARF Distal end of the gynoecium and in replum development [71] BRASSINOSTEROID ENHANCED EXPRESSION1 (BEE1) AT1G18400 bHLH Reproductive tract development [105] BRASSINOSTEROID ENHANCED EXPRESSION3 (BEE3) AT1G73830 bHLH Reproductive tract development [105] BREVIPEDICELLUS (BP) AT4G08150 Homeobox Replum development [104] CUP-SHAPED COTYLEDON 1 (CUC1) AT3G15170 NAC SAM development and organ boundary definition [88,89] CUP-SHAPED COTYLEDON 2 (CUC2) AT5G53950 NAC SAM development and organ boundary definition [88] CRABS CLAW (CRC) AT1G69180 YABBY Gynoecium primordium specification [47] FILAMENTOUS FLOWER (FIL) AT2G45190 YABBY Valve development [101] FRUITFULL (FUL) AT5G60910 MADS-box Valve development [102] HALF FILLED (HAF) AT1G25330 bHLH Reproductive tract development [105] HECATE 1 (HEC1) AT5G67060 bHLH adaxial-identity in gynoecium [61] HECATE 2 (HEC2) AT3G50330 bHLH adaxial-identity in gynoecium [61] HECATE 3 (HEC3) AT5G09750 bHLH adaxial-identity in gynoecium [61] INDEHISCENT (IND) AT4G00120 bHLH Valve margin tissue development and seed dispersal [55,125] NGATHA (NGA) AT2G46870 AP2/B3-like Style specification [64,65] NO TRANSMITTING TRACT (NTT) AT3G57670 C2H2 Transmitting tract development [98−100] PIN-FORMED 1 (PIN1) AT1G73590 PIN Proximal-distal axis establishment [53] PIN-FORMED 3 (PIN3) AT1G70940 PIN Proximal-distal axis establishment [127,128] REPLUMLESS (RPL) AT1G02065 Homeobox CMM development and replum morphogenesis [97] SEPALLATA (SEP) AT5G15800 MADS-box Gynoecium identity [28] SHATTERPROOF 1 (SHP1) AT3G58780 MADS-box Dehiscence zone differentiation [114] SHATTERPROOF 2 (SHP2) AT2G42830 MADS-box Dehiscence zone differentiation [114] SPATULA (SPT) AT4G36930 bHLH Style development [55,84] SEEDSTICK (STK) AT4G09960 MADS-box Funiculus and ovule development [111] STYLISH (STY) AT3G51060 SHI Gynoecium apex development [67] SHOOT MERISTEMLESS (STM) AT1G62360 Homeobox SAM development [94,95] TEOSINTEBRANCHED1 / CYCLOIDEA / PCF15 (TCP15) AT1G69690 TCP Gynoecium apex development [75] YABBY 3 (YAB3) AT4G00180 YABBY Valve development [101] YUCCA 4 (YUC4) AT5G11320 YUC Gynoecium primordium specification and gynoecium apex [66] WUSCHEL (WUS) AT2G17950 Homeobox FMs development [29] Expression of AG in the FM gradually shuts down the WUS expression at stages 4 and 5 through multiple independent pathways[33,36]. First, AG physically interacts with Polycomb Group (PcG) protein TERMINAL FLOWER 2 (TFL2, also known as LIKE HETEROCHROMATIN PROTEIN 1, LHP1) and binds to the 3’ and 5’ regulatory sequences of WUS locus, respectively, resulting in a chromatin loop, which in turn represses WUS expression by decreasing the DNA accessibility for RNA polymerase II[37,38]. Secondly, AG directly activates the expression of C2H2 zinc-finger domain and an EAR motif containing transcription factor, KNUCKLES (KNU)[39]. Meanwhile, AG also indirectly induces the expression of MINI ZINC FINGER 2 (MIF2) (Fig. 3; Table 1)[40]. As a result, in the expression domain of AG, two repressive protein complexes are formed on the WUS locus: (1) KNU recruits transcription co-repressor TOPLESS (TPL) together with HISTONE DEACETYLASE 19 (HDA19) and MIF2 that removes the active acetylation maker on the histones and represses WUS expression (Fig. 3; Table 1)[41]; (2) KNU directly interacts with FERTILIZATION INDEPENDENT ENDOSPERM (FIE), which is a component of Polycomb Repressive Complex 2 (PRC2)[42]. The resultant KNU-FIE-PRC2 protein complex silence the WUS expression by catalyzing trimethylation on lysine 27 of histone H3 (H3K27me3)[37,43]. Taken together, all these factors form a multi-layered regulatory mechanism ensuring that WUS expression is repressed during the gynoecium development.
-
As discussed above, multiple independent pathways are recruited to ensure the balance between gynoecium primordium initiation and stem cell termination, which is essential for further gynoecium development.
Immediately after initiation, the gynoecium starts to grow rapidly along the medial-lateral and adaxial-abaxial axes, during which distinct cell types are differentiated and patterned into tissues. A recent study employed quantitative live imaging techniques showing that the Arabidopsis gynoecium development is determined by two orthogonal, time-shifted differentiation gradients: an early mediolateral gradient controlling valve morphogenesis and a late longitudinal gradient regulating style differentiation[20]. It is proposed that regional growth gradients fine-tune the developmental program of the gynoecium patterning process (Fig. 2a & c)[20]. The rapid growth of the gynoecium primordium is driven by fast cell expansion, which mainly relies on the modification of cell wall components, such as cellulose, pectin, and glycoproteins[44,45]. Auxin in the cells promotes the cell wall loosening and expansion[46]. Indeed, one of the AG-mediated pathways is the upregulation of YUCCA 4 (YUC4) by directly activating the YABBY transcription factor CRABS CLAW (CRC) expression (Fig. 3; Table 1)[47]. YUC4 encodes a flavin monooxygenase involved in auxin biosynthesis[48]. The resultant YUC4 expression in the gynoecium primordium generates an auxin maximum in the abaxial cell layer at the gynoecium apex that facilitates the fast cell expansion (Figs 2a, 3; Table 1)[47−49].
-
In addition to promoting cell elongation, auxin also plays a prominent role in patterning the gynoecium along the proximal-distal axis. In agreement with this notion, blocking polar auxin transport (PAT) by N-1-naphthylphthalamic acid (NPA) or mutations affecting the integrity of the auxin pathway result in the deformation of the tissues along the proximal-distal axis (Fig. 2a, c)[50,51].
During the gynoecium development, the earliest hallmark of auxin signaling is the observation of two lateral foci at the apex of the primordium, which manifests the two carpels (Fig. 2c)[52]. These two auxin maxima are generated by a combination of the apical localization of the auxin efflux carrier PIN-FORMED 1 (PIN1) proteins in the epidermis, which transport auxin from the base to the apex and local auxin biosynthesis by YUC4 (Fig. 3; Table 1)[48,52,53]. The establishment of these two lateral auxin maxima is crucial for the early outgrowth of lateral domains in the gynoecium as NPA treatment retards the lateral development of the gynoecium primordium[19,54]. At stage 10 of gynoecium development, the gynoecium is fused at the apex forming a radially symmetrical structure that allows the differentiation of style and stigmatic tissues (Fig. 1b)[11]. The switch in this symmetry transition is facilitated by the establishment of two additional medial auxin foci and subsequently a ring-formed auxin signal at the gynoecium apex (Fig. 2c)[52,54]. At the genetic level, the establishment of the two medial auxin foci is promoted by two basic helix-loop-helix (bHLH) transcription factors SPATULA (SPT) and INDEHISCENT (IND) (Figs 2b, 3; Table 1). In both spt and ind;spt mutants, the stepwise auxin distribution is blocked at the lateral two-foci stage, the two medial and the following ring-formed auxin distribution failed to establish[52,55]. The resultant fruits from spt or ind;spt mutants develop unfused gynoecia with a cleft in the medial region[52]. At the gynoecium apex, SPT and IND form heterodimers, which in turn repress the expression of AGC3-type protein kinase PINOID (PID)[56]. PID phosphorates PIN proteins that facilitate their polar localization at the plasma membrane (Figs 2b, 3; Table 1)[57]. Due to the repression of PID at the gynoecium apex, PIN proteins become apolarly localized, resulting in the establishment of two additional auxin signaling points at the medial region, which is important for subsequent transformation into a radial ring pattern at the gynoecium apex[52,58,59]. It was recently discovered that the SPT-IND complex activates the expression of core cell-cycle regulators, CYCLIN-D1;1(CYC-D1;1) and CYC-D3;3, thus maintaining cell division orientation perpendicularly to the direction of organ growth in medial-apical cells. This anticlinal (transversal cell division) is required for symmetry transition[60].
In addition to setting up the medial auxin foci, a functional SPT is also indispensable to the formation of ring-formed auxin signaling at the gynoecium apex[61]. During this process, SPT exerts its function in conjunction with a set of bHLH transcription factors, HECATE (HEC1-3), to regulate the expression of HOMEOBOX ARABIDOPSIS THALIANA 3 (HAT3) and ARABIDOPSIS THALIANA HOMEOBOX 4 (ATHB4), thereby mediating the final step in the style radialisation process by establishing the ring-formed auxin pattern (Figs 2b, 3; Table 1)[61]. Interestingly, a recent study revealed that SPT function is modified at the post-translational level by O-glycosylation[62]. The attachment of O-fucose and O-GlcNAc, respectively, to the SPT enhances the binding affinity of SPT to the PID locus[62]. Two O-glycosyltransferase, SPINDLY (SPY) and SECRET AGENT (SEC), synergistically control style morphogenesis by promoting the activity of SPT via O-glycosylation[62]. Therefore, the boundary delineation between ovary and style is defined by the dynamic changes in auxin signaling pattern at the gynoecium apex, which is promoted by apolar PIN locations mediated by SPT and associated factors (Fig. 2b).
The resulting ring-formed auxin maxima at the gynoecium apex promote the further differentiation of apical structures, including the stigma and style (Fig. 2c)[63]. The expression of IND and SPT is sequentially activated by HEC proteins together with the B3 transcription factor NGATHA (NGA)[64,65]. Then SPT-IND interacts with HECs and NAG to form a high-order protein complex to repress PID expression, and therefore induce the apolar localization of PIN proteins to sustain the auxin maxima at the gynoecium apex[65]. Additionally, NGA activates the expression of SHORT NTERNODES/STYLISH (SHI/STY), which encodes a zinc-finger domain-containing transcription factor, and cooperatively functions with STY to induce auxin biosynthesis by activating YUC4 expression (Figs 2b, 3; Table 1)[66−68]. NGA, STY, and YUC4 are expressed in the distal end of the gynoecium, resulting in a local auxin production at the gynoecium apex (Figs 2b, 3; Table 1)[64−66]. The auxin maximum in the style and stigma is disrupted in either spt, hec1;hec2;hec3, or high-order nga mutants[65,68]. In plant cells, auxin signal is translated into transcriptional output by AUXIN RESPONSE FACTOR (ARF) transcription factors[69,70]. ARF3, also known as ETTIN (ETT), is expressed in the distal end of the gynoecium and the replum (Fig. 3; Table 1)[71]. The gynoecium of ett mutant displays strong polarity defects including extended style and gynophore regions, a reduction in the ovary size, and ectopic extension of papillae cells in the style and replum[71]. Interestingly, ETT was found to bind to auxin and interact with a plethora of transcription factors in an auxin-sensitive manner[72−74]. It is therefore suggested that ETT acts as an interpreter of auxin concentration in the apical part of the gynoecium by interacting with specific proteins, leading to the differential downstream transcriptomes in response to auxin signals[73]. However, how ETT interprets the auxin gradient along the distal proximal in delineating the boundaries of stigma, style, and replum remains largely unknown.
Finally, differentiation of style and stigma is a determinate process that is redundantly regulated by TEOSINTE BRANCHED 1/CYCLOIDEA/PCF (TCP) transcription factors[75]. Expression of TCPs is predominantly localized in the apex of the gynoecium (Figs 2b, 3; Table 1)[75]. Gynoecia of high-order tcp mutants develop longer and narrower style[75]. However, when crc or nga mutation was introduced into the high-order tcp mutant, the style was changed into fascinated indeterminate lamellar structures[75]. Like TCP genes, STIGMA AND STYLE STYLIST (SSS) genes are also expressed in the apical part of the gynoecium. SSS encodes angiosperm-specific proteins with unknown function[76]. SSSs act downstream of NGA transcription factors and cooperatively regulate the style and stigma growth via controlling cell elongation (Fig. 3; Table 1)[76].
-
The FM stops producing new organs after the initiation of gynoecium[12,13]. However, new tissues and cell types, e.g., septa, placenta tissues, and ovules continue to differentiate inside the gynoecium, indicating the maintenance or the re-establishment of the meristematic identities within the gynoecium[14,15]. This 'meristematic-like zone' was termed as the carpel margin meristem (CMM), located in the medial domain, which is established along with the differentiation of the medio-lateral axis in early gynoecium development[14,15].
In the SAM, the stem cell identity is maintained by increasing cytokinin levels or sensitivity[77,78]. Reduced cytokinin levels or signaling results in a smaller SAM[78]. Analysis of cytokinin transcriptional response reporter two-component signaling sensor (TCS) has shown that cytokinin signaling is active in the medial domain of the gynoecium primordium, where the potential CMM will be initiated (Fig. 2c)[79]. Later in development, TCS activity is specifically localized in the CMM and the tissues differentiated from the CMM (placental tissues, septa), and the valve margins of the mature gynoecia and young fruits (Fig. 2c)[79]. Therefore, the CMM cells are reminiscent of the stem cells in the SAM in terms of cytokinin signaling properties.
In plants, cytokinins are synthesized by the ISOPENTENYL TRANSFERASE (IPT) and LONELY GUY (LOG) enzymes and metabolically degraded by CYTOKININ OXIDASE (CKX) enzymes[80−82]. The cytokinins are perceived by the ARABIDOPSIS HISTIDINE KINASE (AHKs) receptors, and then the signals are transduced as transcriptional output via phosphorating the B-type ARABIDOPSIS RESPONSE REGULATORS (ARRs)[82,83]. Gynoecium with increased cytokinin levels by exogenous cytokinin treatment or IPT over-expression leads to over proliferation of the medial tissues from the CMM, whilst the gynoecium of arr1;arr10;arr12 mutant has limited capacity of tissue proliferation from the CMM, as evidenced by fewer ovules, a reduction in transmitting tract tissues, and defects in septum fusion (Fig. 3; Table 1)[83,84]. Intriguingly, CKX genes are dynamically expressed in the CMM cells, suggesting cytokinin levels and signaling are strictly fine-tuned in a spatio-temporal manner[85]. Additionally, the CUP-SHAPED COTYLEDON 1 (CUC1) and CUC2 encode paralogous NAC transcription factors that are involved in SAM development and organ boundary definition[86]. Expression of CUC genes is negatively regulated by microRNA164 (miR164) at the post-transcriptional level[87]. In the gynoecium, both CUC1 and CUC2 are specifically expressed in the CMM region (Fig. 3; Table 1)[88]. CMM identity, placenta development and ovule initiation are severely compromised in the cuc1;cuc2 double mutant, whilst plants expressing miR164-resistant forms of CUC1 and CUC2 resulted in extra CMM activities[79,87,88]. In the SAM, CUCs are activated by class I KNOX transcription factor SHOOT MERISTEMLESS (STM) and then CUCs reinforce the STM expression constituting a double positive feedforward loop to sustain the SAM function (Fig. 3; Table 1)[88−91]. STM is known to balance the stem cell proliferation by activating cytokinin biosynthesis while repressing gibberellin activities[92,93]. Interestingly, the expression of STM is specifically localized in the CMM but significantly reduced in the cuc1;cuc2 mutant[94]. STM knock-down by RNAi produces flowers without the gynoecium[95]. Taken together, these observations suggest CUCs promote stem cell maintenance in the CMM by inducing cytokinin biosynthesis through the STM-mediated pathway. This assumption is in agreement with the strong expression of TCS::GFP in the CMM (Fig. 2c)[79]. In addition to CUCs and STM, SPT is also found to be expressed in the CMM and mediates the cytokinin signaling by directly binding to the ARR1 promoter[84,95,96]. In summary, all these studies revealed the critical role of cytokinin in the establishment of the CMM and the development of the middle region of a gynoecium.
The differentiation of distinct tissues along the medio lateral axis is governed by a regulatory pathway with multiple transcription factors involved, many of which are exclusively expressed in distinct domains[14]. In the medial region of a gynoecium, the replum is derived from the CMM[11]. The homeodomain transcription factor, REPLUMLESS (RPL), is expressed in the medial regions and plays a crucial role in CMM development and replum morphogenesis (Figs 2d, 3; Table 1)[97]. The rpl gynoecium exhibits severe defects in the CMM, leading to a reduction in replum size and ovule number, loss of the septum, and carpel fusion defects[97]. Inside the CMM, the differentiation of transmitting tract is governed by NO TRANSMITTING TRACT (NTT) gene[98,99]. Interestingly, the replum is completely lost in the rpl;ntt double mutant, indicating a synergistic development of replum and transmitting tract in the middle region[100]. On the other hand, the establishment of valve identity in the lateral region is defined by the specific expression of two YABBY transcription factors, FILAMENTOUS FLOWER (FIL) and YABBY 3 (YAB3), in the valves (Figs 2d, 3; Table 1)[101]. The fil;yab3 double mutant develops gynoecia with mis-patterned valve identities[101]. FIL and YAB3 affect valve development partially by activating the expression of the MADS-box transcription factor, FRUITFULL (FUL)[102]. The valves of ful mutant are deformed and failed to grow after pollination[101,102] (Discuss below). Another important function of FUL is to repress the expression of RPL in the valve cells and restrict its expression in the medial region (Figs 2d, 3; Table 1)[100,103]. The expression of RPL in the CMM is positively regulated by a class I KNOX homeodomain transcription factor BREVIPEDICELLUS (BP) (Figs 2d, 3; Table 1)[100,101,104]. BP is activated by NTT in the CMM and then physically interacts with RPL to form a heterodimer that restricts the YAB gene expression to the lateral region[99−101,104]. Therefore, the gynoecium patterning along the medio lateral axis depends on both the maintenance of CMM activity and the antagonistic action of valve and replum identity genes, respectively (Figs 2d & 3).
-
In Arabidopsis, the gynoecium starts to grow into a fruit after pollination[11]. The directional growth of the pollen tube from the stigma to the unfertilized ovules is facilitated by the development of the transmitting tract. Three closely related bHLH transcription factors, HALF FILLED (HAF), BRASSINOSTEROID ENHANCED EXPRESSION1 (BEE1) and BEE3, redundantly regulated the transmitting tract development by promoting cell death. The expression of these genes is localized in the transmitting tract and depends on NTT and auxin signaling pathways (Fig. 3; Table 1)[105]. After pollination, the fruit growth is manifested by the dramatic elongation along the proximal-distal axis, while in the medio lateral axis is less pronounced (Fig. 1b & d). A recent study combined live imaging with genetic analysis showed that fruit elongation is triggered by a mobile signal generated from the fertilized ovules at stage 13[106]. This active fruit growth in the post-pollination stages is largely driven by extensive cell expansions with little if any contribution from cell division[107]. The fruit morphogenesis program is different from that observed in leaves, sepals, or roots, in which a spatial-temporal dynamic change in cell division and expansion is involved[107,108]. In this regard, even if the fruits have evolved from the modified leaves, the genetic pathway underlying their morphogenesis process has been modified to optimize the fruit development process triggered by the signal from fertilization.
As discussed above, the MADS-box transcription factor FUL is a master regulator of valve identities[102]. The FUL gene is strongly expressed in the inflorescence meristem, the floral shoot apex, and then specifically in the ovary walls and fruit valves[102]. In the valve cells, FUL integrates the auxin signals by forming heterodimers with ARF6 or ARF8. The FUL-ARF6/8 complexes in turn activate the miR172C gene expression in the valves[109]. miR172C negatively targets the mRNA of APETALA 2 (AP2), which is a transcriptional repressor (Fig. 2d & e)[109,110]. In this way, FUL promotes fruit valve growth by eliminating the negative effect of AP2 in the valves (Fig. 3; Table 1)[109,110]. In addition to the FUL-ARF6/8 mediated auxin pathway, cytokinin was recently demonstrated to have a negative effect on fruit elongation[111]. The cytokinin degrading enzyme, CYTOKININ OXIDASE/DEHYDROGENASE 7 (CKX7), is actively expressed in the valves. Expression of CKX7 is directly activated by the MADS-box transcription factor SEEDSTICK (STK) (Fig. 2d)[110]. In agreement with this regulatory relationship, both the ckx7 and stk mutants produce short fruits, as expected from the excessive accumulation of cytokinins[111]. CKX7 is activated by the STK, which is also responsible for ovule and funiculus development (Fig. 3; Table 1)[112]. Interestingly, the expression of FUL is also found to depend on the STK, indicating a crosstalk between auxin and cytokinin’s in fruit elongation (Fig. 2d)[111]. However, the STK is primarily expressed in the funiculus and ovules, where it regulates funiculus development and ovule patterning[111,112]. Whether the shorter fruits in the stk result from the disruption of cytokinin degradation or defects in ovule development remains to be clarified soon (Figs 2d, e & 3).
-
The final developmental event of a fruit is to shatter the pod, release the seeds, and ultimately promote seed dispersal[11,113]. Fruit dehiscence and seed abscission depend on a similar mechanism with a few specialized cells in the dehiscent zone (DZ)[111,113,114]. For separation to take place, mechanical forces generated in the surrounding tissues or external agents trigger the detachment of cells at the separation layer (SL) (Fig. 2e)[115]. The DZs in the Arabidopsis silique are located at the very edge of the valve and adjacent to the replum. In a cross-section of a mature Arabidopsis fruit, the DZ consists of two distinct cell types, (1) the SL: 1−2 layer of isodiametric parenchyma cells that are positioned adjacent to the replum[11]; and (2) the lignified layer (LL), a single cell layer with rigid secondary wall thickening that relates to the lignified enb layer of the valve (Fig. 2e)[116]. When the fruits mature, the shrinkage of the parenchyma cells in the valve generates a mechanical force that promotes the valve to release the force at the weakest point, the separation layer, and hence, the pod dehiscence occurs (Fig. 2e & f).
In Arabidopsis, the specification of the DZ is governed by a delicate genetic regulatory network with multiple transcription factors and hormones involved[113]. This topic has recently been extensively reviewed[113]. In brief, the MADS-box transcription factors SHATTERPROOF 1 and 2 (SHP1/2) and IND determine both SL and LL cell identities, whilst the SL layer differentiation is additionally controlled by a myb/bHLH transcription factor ALCATRAZ (ALC) (Figs 2e, 3; Table 1)[114−117]. All these four transcription factors are specifically expressed in the developing DZ, with SHP1/2 redundantly activating IND and ALC expression[114−117]. These DZ identity genes are excluded from the replum and valve by RPL and FUL, respectively (Fig. 2e)[97,102,114,118]. In either ful or rpl mutants, the LL cell identity is ectopically extended into the replum and valve regions[97,102,116]. The ectopic lignification of the parenchyma cells in the valves partially explains why the ful fruits fail to elongate after pollination[102]. Interestingly, over-expression of NTT disrupts the valve margin and lignified enb layer development by suppressing the FUL expression, indicating a regulatory relationship between NTT and RPL[119]. In addition, AP2 was recently discovered as a negative regulator of both replum and DZ identities by fine-tuning the expression level of SHP1/2 and IND in the DZ and RPL in the replum, respectively[120]. The output of the DZ identity genes is the differentiation of distinct cell types by activating different regulatory cascades. The development of the secondary cell wall in the LL is controlled by the NAC transcription factor NAC SECONDARY WALL THICKENING PROMOTING FACTOR1 (NST1) (Figs 2e, 3; Table 1)[121−123]. NST1 is a master regulator of cell wall thickening by activating genes involved in lignin and cellulose synthesis[122]. NST1 acts downstream of IND in specifying the LL cell identities, and expression of NST1 is evident in both the LL and the valve enb layer[123]. The nst1 fruits are indehiscent due to defects in the secondary cell wall thickening in both LL and enb cells[123]. Meanwhile, the breakdown of the cell-extracellular matrix in the SL cells facilitates the fruit dehiscence process[124]. The ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE 1 (ADPG1) and ADPG2 encode POLYGALACTURONASE (PG) that are necessary for the degradation of the cell wall matrix. Both ADPG1 and ADPG2 are positively regulated by IND in the DZ region, where they are required to trigger cell separation and fruit dehiscence at maturity (Figs 2e, 3; Table 1)[124].
On top of transcription factors, phytohormones also have been elucidated to play important roles in specifying the DZ identities. During fruit development, cytokinin signal reporter (TCS::GFP) is specifically active in the DZ, and exogenous cytokinin (benzylaminopurine, BAP) application restored the indehiscent fruit defect in both ind and shp1;shp2 double mutant (Fig. 2c)[79]. Moreover, the TCS::GFP expression is absent in the DZs of either ind or shp1;shp2 double mutants[79]. It is, therefore, likely that one of the downstream events of SHP-IND module is to generate a cytokinin maximum in the DZ. However, strong expression of CKX7 in the fruit valve under the control of the STK-FUL module could also potentially restrict the cytokinin to the DZs. It would be interesting to test if the cytokinin maximum in the DZ is generated by local biosynthesis or suppression of degradation by the SHP-IND module in the DZ[79,125]. In addition to cytokinins, IND also regulates the PIN localization by directly activating PID and WAG2[126,127]. Consequently, a dynamic auxin signaling pattern is established in the DZ in a stage-specific manner to ensure the precise differentiation of DZs. At stages 14−16, when the LL and SL start to differentiate via asymmetric cell divisions, IND represses the expression of WAG2 and PID (Fig. 2e)[125]. Because of this IND-mediated repression of WAG2 and PID, PIN3 plasma membrane abundance is decreased, which in turn causes influxes of auxin from the surrounding tissues and generates an auxin maximum in the DZ[125]. This auxin maximum is required to promote the asymmetric cell divisions characterizing the early dehiscence zone patterning[125]. At stage 17, when the fruit reaches its full length with cells in the DZ finalizing their differentiation, IND directly activates WAG2 while continuing to repress PID (Fig. 2e)[127]. Upregulation of WAG2 shifts PIN3 to the lateral side of the DZ cells that efflux auxin out and generate auxin minimum in the DZ[127]. A recent mathematical modeling study further revealed that the 'flux-passage' auxin flux pattern causes auxin minimum in the DZ, substantiating the role of apolar localization of PIN3 in DZ cells[128]. Both the auxin maxima and minimum are required for the DZ differentiation as fluctuation in auxin concentration by ectopically expressing the AGC3 protein kinase (WAG2 or PID) or auxin synthetic gene (iaaM) disrupts DZ formation and abolishes fruit dehiscence[125,127] Finally, IND also mediates gibberellin (GA) accumulation by directly activating the expression of GA biosynthetic enzyme GA3ox (Fig. 2e)[129]. High levels of GA in the DZ destabilizes DELLA repressors and frees ALC from DELLA-mediated repression in SL development[129]. In conclusion, the DZ development entails cooperative interactions between the SHP-IND module and associated phytohormonal networks. Cytokinin and auxin signaling are overlapped and regulated by IND in the early patterning stage of the DZ[79,125]. How these two hormones are intercrossed to direct the DZ differentiation warrants further investigation.
Like the pod dehiscence, the seed abscission from the funiculus depends on the development of the seed abscission zone (SAZ) in the distal end of the funiculus (Fig. 2f). The SAZ is composed of a single LL in a ring pattern surrounding the vascular bundle and an SL with less lignified cells at the edge of the funiculus (Fig. 2f)[130]. STK interacts with the SEUSS (SEU) co-repressor to down-regulate the HEC3 expression in the LL. The seeds fail to abscise in the stk mutant due to ectopic expression of HEC3 and lignification of the funiculus[130]. Since HEC3 is a close homolog of IND, while STK is closely related to SHPs, it is therefore suggested that genetic networks regulating pod dehiscence and seed abscission are highly conserved[130]. However, this conclusion awaits further substantiation by elucidating the role of hormones in SAZ differentiation (Fig. 2e, f).
-
In the past decades, our understanding of the genetic regulation of gynoecium patterning and fruit development has been greatly advanced by the continuous effort to dissect the genetic components involved in this process. We are also starting to appreciate the role of plant hormones, in particular auxin and cytokinin, in the robust GRN balancing cell proliferation and differentiation during gynoecium and fruit development. Nonetheless, some important questions remain to be addressed in the near future. For example, gibberellins are known to be important hormones in stem cell maintenance and fruit growth, however, it is unclear how gibberellin signals are rewired with CK in CMM development and fruit growth. Analyzing the gibberellin signaling patterns using the recently developed ratiometric GA signaling biosensor would help to clarify this question[131]. Additionally, the development of the papillae cells at the top of the gynoecium is unique in angiosperms, which could be related to speciation as they are the front line to determine if the plants are competent to outcross or not. The differentiation of stigmas is associated with maxima at the gynoecium apex. How this auxin maximum is sensed and translated into the developmental program directing the stigma and style differentiation is a very intriguing topic that warrants further in-depth investigation. Finally, in addition to the cylindrical fruit shape of Arabidopsis, the Brassicaceae exhibit a great variation in fruit shapes, which are adaptive to specific dispersal strategies. Recent studies have shown that while most of the genetic components have been largely evolutionary conserved in different species, despite the fact that some of them have been diversified in terms of both downstream genes and expression patterns[132,133]. Future comparative genomic and functional analysis in closely related Brassicaceae species with distinct fruit shapes will test the conservation of the genetic pathway underlying fruit development, by which the genetic targets could be identified to design better-performed crops using CRISPR and other modern genetic tools.
We apologize to authors whose work were unable to discuss in this review owing to space constraints. We are grateful to Dr Tian-Feng Lv, Zhi-Cheng Hu, Yao Zhang, Jeonghwan Ahn, Quan Yuan, Hao-Ran Sun, Xiao-Yu Chen, and Fan-Ding Zhu for critically reading the manuscript and providing comments prior to submission. Research in the Dong Lab was supported by grants from the National Key Research and Development Program of China (2022YFF1301704), the National Natural Science Foundation of China (32170227 and 32221001) and grants from the Chinese Academy of Sciences.
-
The authors confirm contribution to the paper as follows: study conception and design: Ding Y, Dong Y; figure preparation, draft manuscript preparation: Ding Y; writing - sections on Arabidopsis gynoecium: Gao F; manuscript revise: Li X, Dong Y. All authors reviewed the results and approved the final version of the manuscript.
-
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
-
The authors declare that they have no conflict of interest.
- Copyright: © 2024 by the author(s). Published by Maximum Academic Press on behalf of Hainan Yazhou Bay Seed Laboratory. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Ding YN, Gao F, Li XR, Dong Y. 2024. Genetics and molecular regulation of gynoecium patterning and fruit development in Arabidopsis. Seed Biology 3: e018 doi: 10.48130/seedbio-0024-0016
Genetics and molecular regulation of gynoecium patterning and fruit development in Arabidopsis
- Received: 16 August 2024
- Revised: 29 September 2024
- Accepted: 21 October 2024
- Published online: 22 November 2024
Abstract: The evolutionary origin of fruits represents a key morphological innovation associated with angiosperm diversification by promoting seed dispersal. Fruits are also a nutritional source for the human diet. The fruit is derived from the gynoecium in the strictest sense, the female reproduction produced at the center of a flower. Fruit development and gynoecium patterning are interconnected processes requiring consecutive differentiation of multiple distinct tissues and rapid cell expansion after pollination. In the past decades, our understanding of fruit development has been extensively extended by active studies of Arabidopsis. More importantly, the roles of phytohormones in directing tissue differentiation along the polarity axes during gynoecium patterning and fruit development have just begun to be recognized. In this review, a comprehensive summary of the latest advancements in the gene regulatory networks (GRNs) that control tissue differentiation during gynoecium and fruit development in Arabidopsis are provided. We also discuss hormonal crosstalk and associated pathways into the GRNs by showing examples of how hormones could modify GRNs in fruit development. The unresolved questions concerning the fruit development are further highlighted, with a particular focus on the evolution of fruit, which opens the avenue for further studies.













