-
Tomato (Solanum lycopersicum) is affected by several soil-borne diseases, however, bacterial wilt disease caused by Ralstonia solanacearum is of great concern to farmers due to the pathogen's high genetic diversity, wide host range, and adaptation to several environmental conditions[1]. The destructive nature of the disease in vegetable production and more especially on the economy of tomatoes has been reported by multiple studies. Previous studies[2−4] have reported a high yield loss of about 90% in tomatoes under severe bacterial wilt disease outbreaks. Since the wilt disease on tomatoes was reported in Ghana[5], no resistant cultivar has been identified, confirming that most commercial tomato varieties are susceptible to the disease[6].
High pathogen aggressiveness, numerous host ranges, and highly favorable environmental conditions make controlling of the disease difficult. Notwithstanding, several cultural practices including field sanitation, soil amendments, and field and crop rotations have been employed to reduce the impact of the disease. In addition to the limited number of effective chemical management strategies, the use of synthetic chemicals is highly discouraged due to numerous harmful side effects on the environment, animal and human life, and increased development of chemical resistance[7,8]. The habitation of the pathogen in the xylem of its host and the ability to reside deep in the soil makes the use of chemicals ineffective[9].
The use of resistant varieties to manage diseases is both environmentally friendly and relatively cost-effective for farmers[10]. A major hindrance to the use of resistant varieties in tomato production has been the negative correlation between crop yield and bacterial wilt disease resistance[11]. To overcome this, the grafting of desirable susceptible varieties onto resistant rootstocks has recently been explored and popularized as an effective control mechanism against R. solanacearum-induced wilting in several crops. Earlier studies[12,13] have reported on the use of resistant solanum rootstocks to reduce the incidence and severity of bacterial wilt disease in susceptible tomatoes culminating in the high demand for bacterial wilt disease suppressive rootstocks in several countries including the USA. In addition to disease management, utilization of rootstocks has been found to improve plant establishment in disease-endemic fields, increase the tolerance level of the scion to environmental stress, and ultimately increase the yield of the desired crop[12,14]. Given the above benefits, multiple efforts have been geared toward screening and identifying R. solanacearum-tolerant rootstocks. For example, Ramesh et al. & Namisy et al.[3,9] screened and identified varieties of Solanum torvum rootstocks resistant to bacterial wilt disease. Again, in several countries, commercial rootstocks have been developed and marketed to farmers to control the disease. In Ghana, however, no commercially available rootstock has been developed and little information exists on the use of rootstocks to manage bacterial wilt disease in tomatoes. Equally, farmers do not have access to any management intervention against the wilt disease, hence leaving infected plants unmanaged. Developing an accessible technology that is eco-friendly, easy to adapt, and less costly to smallholder farmers will enormously increase tomato production and productivity in the country. Identifying and promoting the use of resistant rootstocks could be a valuable tool for tomato growers in Ghana. To achieve this, there is the need to continually screen available germplasm or accession to identify promising lines or rootstocks. Therefore, in this study, it was hypothesized that evaluating some solanum plants may lead to the identification of R. solanacearum-resistant accession(s) for use as rootstock. To test this hypothesis, 13 solanum accessions were evaluated to select resistant rootstocks to provide alternate control and mitigate the negative impact of bacterial wilt disease in Ghana.
-
Thirteen accessions comprising seven tomatoes (Solanum lycopersicum), and six garden eggs (Solanum melongena) breeding lines and cultivars were obtained from different sources (Table 1). Seeds of the various accessions were surface sterilized in 70% ethanol and serially rinsed in sterile distilled water as described by Davoudpour et al.[15] before sowing in separate trays filled with commercial cocopeat.
Table 1. List of tomato and garden egg accessions evaluated in the study.
No. Code Solanum spp. Biological status Source Country/Region of origin 1 BL 729 Tomato Breeding line Worldveg Taiwan 2 BL 9884 Tomato Breeding line Worldveg Taiwan 3 L_020 Tomato Open pollinated TGRC, UC Davis USA 4 GC Tomato Open-pollinated TGRC, UC Davis USA 5 BL 1534 Tomato Breeding line Worldveg Taiwan 6 GD Tomato Open pollinated TGRC, UC Davis USA 7 GG Tomato Open pollinated TGRC, UC Davis USA 8 CRI-06 Garden eggs Breeding line CSIR-CRI Ghana 9 CRI-04 Garden eggs Breeding line CSIR-CRI Ghana 10 CRI-03 Garden eggs Breeding line CSIR-CRI Ghana 11 CRI-02 Garden eggs Breeding line CSIR-CRI Ghana 12 CRI-01 Garden eggs Breeding line CSIR-CRI Ghana 13 Black Beauty Garden eggs Open pollinated CSIR-CRI Ghana Isolation of R. solanacearum and preparation of bacterial inoculum
-
Pure cultures of the bacterial pathogen were obtained from the Plant Pathology Laboratory of the CSIR-Crops Research Institute (CSIR-CRI), Kumasi, Ghana. The pathogen was initially isolated from garden eggs and tomatoes and properly identified and reported by Newton et al.[16] as R. solanacearum. The obtained pathogen was multiplied on a susceptible tomato variety, Pectomech[16]. To diagnose the presence of the bacterial pathogen, the streaming test was carried out on inoculated plants showing symptoms of wilt disease. The bacterial pathogen was further re-isolated from the infected tomato by dipping a sterilized inoculation loop in the ooze and streaking on Nutrient Agar media[17]. The streaked plates were incubated for 48 h at a temperature of 28 ± 2 °C. Colonies of the bacteria growing on the plates were harvested into sterilized distilled water in glass vials and stored at room temperature until used as inoculum. The inoculum concentration was adjusted to 108 cfu/mL before use.
Phenotypic screening of Solanum plants for R. solanacearum resistance
-
Three-week-old seedlings of each accession were transplanted at a spacing of 0.5 m × 0.5 m, onto raised beds with plot sizes of 10 m × 1 m. Before transplanting, the roots of the seedlings were gently wounded by cutting the tips of the tertiary roots using sterilized scissors. The scissors were sterilized after each use. The wounded roots were dipped separately into R. solanacearum inoculum suspension for 30 min before transplanting. The experiment was laid out in a completely randomized block design with four replications where each replication consisted of 20 plants. The experimental field had initially been cropped to maize and had no history of bacterial wilt disease incidence. All agronomic practices such as the application of fertilizer at the rate of 60-40-40 kg/ha, N, P2O5, and K2O respectively, and weed control were carried out when needed.
Bacterial wilt disease assessment
-
The inoculated plants were monitored daily after inoculation for the appearance of wilt symptoms. Following the expression of wilt symptoms, disease incidence, and severity were recorded every 5 d over 30 d. Disease incidence was determined as the proportion of plants showing wilt symptoms in relation to the number of stands per accession. Plants showing symptoms of bacterial wilt disease were further assessed and scored for disease severity on a 0–5, rating scale (0- no wilted leaves, 5-dead plants) as shown in Fig. 1[9].

Figure 1.
Bacterial wilt disease rating scale (0 = no symptoms, 1 = only one leaf partially wilted, 2 = two or three leaves wilted, 3 = all leaves except two or three wilted, 4 = all leaves wilted, 5 = dead plant).
The disease severity index (DI) was calculated following the formula: DI = (N1 × 1 + N2 × 2 + N3 × 3 + N4 × 4 + N5 × 5)/ (Nt/5)[9], where N1 to N5 = the number of plants with disease rating scale values from 0 to 5, and Nt = the total number of plants observed. Based on the disease index, each Solanum line was categorized as resistant or susceptible as shown in Table 2[18].
Table 2. Scale based on disease index for the classification of solanum germplasm.[2]
DI score (0−1) Reaction 0.00−0.20 Highly resistant 0.21−0.30 Resistant 0.31−0.40 Moderately resistant 0.41−0.50 Moderately susceptible 0.51−0.60 Susceptible 0.61−0.90 Highly susceptible 0.91−1.00 Extremely susceptible Molecular screening of Solanum plants for R. solanacearum resistance
Sampling for genomic DNA extraction
-
Leaves of approximately 0.2 g were collected into sampling bags, transferred into pre-frozen mortars, and homogenized. Subsequently, samples were transferred into 2 mL Eppendorf tubes for DNA isolation.
Extraction of Genomic Deoxyribonucleic Acid (gDNA) and Polymerase Chain Reaction (PCR)
-
Genomic DNA was isolated using CTAB (Cetyltrimethylammonium bromide)[19]. DNA was quantified using a Nanodrop 2000 C Spectrophotometer (Thermoscientific, USA) and quality was checked on a 0.8% agarose gel. Nine bacterial wilt trait-linked markers were used for the study as presented in Table 3[20]. These primers are linked to QTLs (Quantitative Trait Loci) Bwr 12 and 6 which confer resistance to bacterial wilt disease PCR was performed using SeeAmp (Hangzhou Bioer Technology Co. Ltd, China) thermal cycler. The PCR amplification reaction of 10 μL contained 10X DreamTaq PCR buffer, 10 mM dNTPs, 10 μM of forward and reverse primer, 2.5 U/μL DNA polymerase, 50 ng DNA template, and nuclease-free water. For the PCR, three controls were used to prevent the scoring of false bands. These comprised a known positive control, a known negative control, and a no template control (NTC). All samples including the positive controls were duplicated to ensure the reliability and reproducibility of results. Amplified products were separated on 1.5% agarose gel in TBE buffer, stained with ethidium bromide, and an image was captured using AlphaImager HP (Proteinsimple, USA). Scoring of bands was conducted using AlphaImager HP Software Version. 3.4.
Table 3. Sequences and expected product size of primers used for the study.
Trait Primer R-gene Forward primer (5'-3') Reverse primer (3'-5') Annealing temperature (°C) Product size (bp) Ref. Resistant genotype Susceptible genotype Bacteria wilt resistance SLM12–2 Bwr-12 ATCTCATTCAACGCACACCA AACGGTGGAAACTATTG
AAAGG55 209 No reference band [12] SLM12–10 Bwr-12 ACCGCCCTAGCCATAAAGAC TGCGTCGAAAATAGTTGCAT 242 SLM6–124 Bwr-6 CATGGGTTAGCAGATGATT
CAAGCTAGGTTATTGGGCCAGAA 292 SLM6–118 Bwr-6 TCCCAAAGTGCAATAGG
ACACACATAACATGGAGTTCGACAGA 183 SLM6–119 Bwr-6 GCCTGCCCTACAACAAC
ATTCGACATCAAACCTATGAC
TGGA255 SLM6–136 Bwr-6 CCAGGCCACATAGAACTC
AAGACAGGTCTCCATACGGCATC 290 SLM6–17 Bwr-6 TCCTTCAAATCTCCCA
TCAAACGAGCAATTGCAAGG
AAAA186 SLM6–94 Bwr-6 CTAAATTTAAATGGACAA
GTAATAGCCCACGATAGGTTGGTATTTTCTGG 276 Scoring of bands/amplicons
-
The band size for resistant genotypes was scored as present (+) whilst that of susceptible genotypes was scored as absent (−).
Statistical analysis
-
One-way ANOVA at a probability level of 5% (p < 0.05) was performed for the wilting incidence and disease severity index using Statistix version 8.0. Before analysis, data on percent wilting was arcsine transformed to improve normality. Differences between the means were compared and separated using Fisher's Least Significant Difference (LSD) test.
-
The various accessions screened showed symptoms of bacterial wilt disease with varied levels of reaction to the R. solanacearum infection at the various assessment periods. Thirty days after inoculation, significant differences (p < 0.05) in percent wilting incidence were observed among the garden egg accessions. Among the garden egg accessions, wilting incidence ranged from 54.0% to 71.0% in accessions CRI-01 and CRI-03 respectively (Fig. 2). A similar trend was observed among the tomato accessions evaluated as significant differences (p < 0.05) in disease incidence were observed among them. Among the tomato lines, accessions L_020, and GD, GC recorded the lowest and highest disease incidence of 56% and 90% respectively (Fig. 2) at the end of the assessment period. Generally, the tomato accessions recorded higher disease incidence compared to garden eggs.
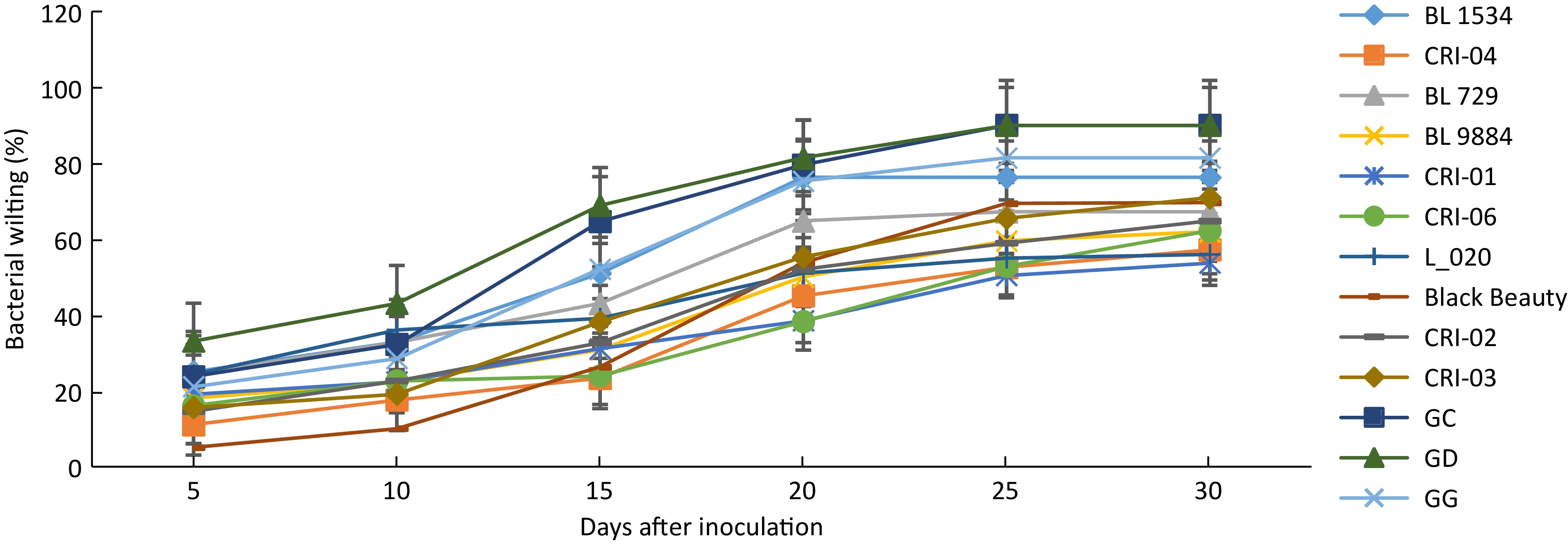
Figure 2.
Wilting incidence among 13 solanum accessions. Error bars represent the standard error of the treatment means.
Disease severity among the garden egg accessions ranged from 0.45 to 0.61 with no significant differences among them (Table 4). This was, however, not the case with the tomato accessions as significant differences (p < 0.05) in disease severity index were observed among the various genotypes. The lowest disease severity (0.37) was recorded for accession (L_020) compared to 0.90 recorded for accession GC (Table 5). Based on the disease severity index, none of the accessions evaluated was found to be highly resistant to the bacterial pathogen. Two accessions (CRI-04 and L_020), however, were moderately resistant with accessions BL1534, GD, GG, and GC recording disease severities above 0.6 and therefore classified as highly susceptible to the pathogen.
Table 4. Mean bacterial wilt disease severity among garden egg accessions evaluated.
Accession Mean disease
severity index (0–1)Host reaction CRI-06 0.45 Moderately susceptible CRI-03 0.56 Susceptible CRI-04 0.37 Moderately resistant CRI-02 0.47 Moderately susceptible CRI-01 0.52 Susceptible Black Beauty 0.61 Susceptible p < 0.05 NS Table 5. Bacterial wilt disease severity and host reaction status among tomato accessions evaluated.
Accession Disease severity index (0–1) Host reaction BL 729 0.53ab Susceptible BL 9884 0.44b Moderately susceptible GG 0.71ab Highly susceptible L_020 0.39b Moderately resistant GC 0.90a Highly susceptible BL 1534 0.70ab Highly susceptible GD 0.70ab Highly susceptible Means followed by different letters are significantly different. Molecular identification of genotypes with BW resistance
-
All the samples produced visible bands following the PCR amplification (Fig. 3), however only samples with the expected band size were scored as positive. Three of the tomato genotypes (L_020, GG, and GC) representing 23%, showed expected bands for all two primers of Bwr-12 whilst none of the garden egg genotypes showed expected bands for Bwr-12 (Table 6). Genotypes that showed expected bands for primers linked to Bwr-6 ranged from two to eight. Four tomato genotypes (BL1534, BL729, GG, and BL9884) and four garden egg genotypes (Black Beauty, CRI-01, CRI-02, and CRI-03) scored the maximum number of alleles for primer SLM 6-118 whilst only two tomato genotypes (BL729 and GD) scored the minimum number of alleles for SLM 6-17. Two of the garden egg genotypes (CRI-01 and CRI-06) showed alleles for only one of the Bwr-6 genes (SLM 6-118 and 6-110 respectively). The genotypes screened for all the nine primers showed alleles ranging from one to seven. Across all the nine primers used, only one tomato genotype (GG) had alleles for seven of the primers whilst none of the garden eggs had alleles across all nine primers. Only one tomato genotype (BL729) had alleles for six of the primers linked to Bwr-6 whilst the garden egg genotype (Black Beauty) had four alleles (Table 6). Genotypes with both Bwr-6 and Bwr-12 exhibit stable resistance against Phylotype I and II strains of the bacteria; hence this study identified some genotypes that had partial resistance (only Bwr-6) and others with durable resistance (combination of both genes). In effect, genotypes that showed amplification for at least one primer of both QTLs (Bwr-6 and Bwr-12) were classified as durable resistance, hence a total of three genotypes were identified. Genotypes with only one marker (either Bwr-6 or Bwr-12) were classified as partial resistance, hence a total of 10 genotypes were identified.
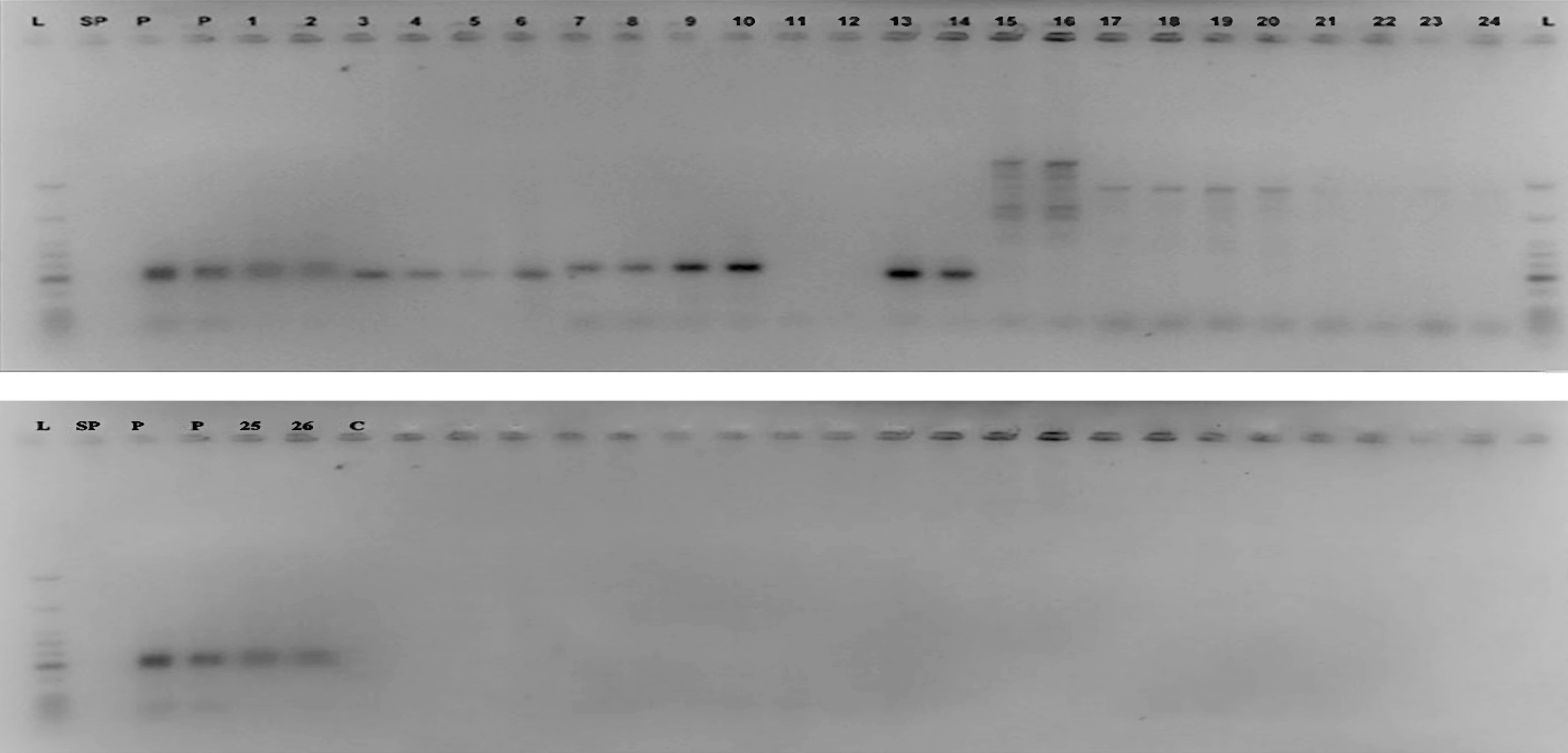
Figure 3.
Agarose gel image of the marker SLM 12-2 for the detection of the Bwr-12 gene L = Molecular weight ladder; SP = Space; P = Positive control; Well 1 & 2 = L_020; 3 & 4 = BL1534; 5 & 6 = BL729; 7 & 8 = GG; 9 & 10 = GC; 11 & 12 = GD; 13 & 14 = BL9884; 15 & 16 = Black Beauty; 17 & 18 = CRI 01; 19 & 20 = CRI 02; 21 & 22 = CRI 03; 23 & 24 = CRI 04; 25 & 26 = CRI 06; C = Negative control.
Table 6. Scores for bacteria wilt resistant gene(s) in tomato and garden egg genotypes.
Genotypes SSR markers Disease reaction Bwr-12 Bwr-6 SLM 12-2 SLM 12-10 SLM 6-136 SLM 6-119 SLM 6-94 SLM 6-118 SLM 6-110 SLM 6-124 SLM 6-17 L_020 +/+ +/+ +/+ +/+ +/+ −/− −/− −/− −/− Durable resistance BL1534 −/− −/− +/+ −/− −/− +/+ −/− −/− −/− Partial resistance BL729 −/− −/− −/− +/+ +/+ +/+ +/+ +/+ +/+ Partial resistance GG +/+ +/+ +/+ +/+ −/− +/+ +/+ +/+ −/− Durable resistance GC +/+ +/+ +/+ −/− +/+ −/− +/+ +/+ −/− Durable resistance GD −/− −/− −/− −/− −/− −/− −/− −/− +/+ Partial resistance BL9884 −/− −/− +/+ +/+ −/− +/+ −/− +/+ −/− Partial resistance Black Beauty −/− −/− −/− +/+ +/+ +/+ +/+ +/+ −/− Partial resistance CRI 01 −/− −/− −/− −/− −/− +/+ −/− −/− −/− Partial resistance CRI 02 −/− −/− +/+ −/− −/− +/+ −/− −/− −/− Partial resistance CRI 03 −/− −/− +/+ −/− −/− +/+ −/− −/− −/− Partial resistance CRI 04 −/− −/− −/− −/− −/− −/− −/− −/− −/− Partial resistance CRI 06 −/− −/− −/− −/− −/− −/− +/+ −/− −/− Partial resistance -
Bacterial wilt disease caused by R. solanacearum is a major constraint to global tomato production[2−4]. The negative effect of the disease is further aggravated by the limited number of management options available to farmers to effectively control the disease[6]. Grafting of susceptible cultivars with desired traits unto a bacterial wilt-resistant rootstock has been identified and promoted as a sustainable means to manage the disease. Both phenotypic and molecular tools have been employed and used successfully to select suitable rootstocks for grafting. The current study screened six tomato and seven garden egg accessions using artificial inoculation procedures and molecular markers to identify and/or select bacterial wilt-resistant rootstocks. The present results showed that all the test materials artificially inoculated with R. solanacearum showed symptoms of wilting. Previous studies[16,21,22] reported that host plants of Solanum spp. present symptoms of wilting following infection and establishment of the pathogen in them. Although all the accessions showed symptoms of infection, they varied in the disease parameters like the incidence and severity of wilting assessed. This assertion is supported by the fact that 57.0% and 16.0% of the tomato and garden eggs lines respectively recorded a wilting incidence of more than 70%. The significant variations in disease expression among and between the accessions as recorded in this study corroborate previous studies[6,23] that reported similar trends in solanaceous cultivars. Based on the phenotypic parameters (wilting incidence and severity scores) measured for this study, none of the test materials screened could be described as resistant to the bacterial wilt disease, although, accessions, L_020 (tomato) and CRI-04 (garden eggs) were found to be moderately resistant. The identification of moderately resistant accessions in the present work supports the findings of Namisy et al. & Stella et al.[9,24] who identified moderately resistant tomato and garden egg lines from field trials for use as rootstocks to manage bacterial wilt disease. The large number of R. solanacearum-susceptible accessions recorded in this study confirms the wide host range of the pathogen causing bacteria wilt disease and the limitation in the identification and selection of resistant rootstocks to manage the disease. Although artificial inoculation procedures have successfully been used to select bacterial wilt-resistant rootstocks, complementing it with marker-assisted selection is considered a standard approach for resistance screening. For this study, in addition to the field evaluation, molecular markers were used to enable efficient identification of genotypes with resistant genes for bacterial wilt diseases as well as multiple resistant gene combinations, which would not have been possible with symptom expression alone. These multiple resistant genotypes could be used in areas where the diseases occur sequentially or simultaneously. Two strains of R. solanacearum that cause bacterial wilt have been reported as being prevalent in Ghana[5]. Bwr-6 and Bwr-12 QTLs have been identified to confer resistance to the disease with Bwr-6 conferring resistance against phylotypes I and II; Bwr-12 conferring resistance against only phylotype I[25]. It has also been reported that genotypes with both Bwr-6 and Bwr-12 show stable resistance against Phylotypes I and II[25], hence any genotype with a combination of both QTLs expresses more durable resistance than genotypes with only one of the genes. Markers used in this study were able to identify genotypes with either Bwr-6 or Bwr-12 or a combination of both. Since genotypes with both Bwr-6 and Bwr-12 show stable resistance, this study identified three tomato genotypes (L_020, GG and GC) that had a combination of both genes and thus, agree with the findings of Carmeille et al.[26] who detected both QTLs in some tomato lines. Furthermore, four of the genotypes had either of the QTLs, indicating partial resistance. All the garden egg genotypes had one or more of the alleles for Bwr-6, indicating partial resistance. In a similar study,[27] it was found that stable QTLs for bacterial wilt resistance in garden eggs were located on chromosomes 3 and 6. The QTLs on chromosome 6 overlap with the BW-resistant QTL (Bwr-6) in tomatoes. This explains the current results where there was no amplification for Bwr-12 QTL in the garden egg genotypes but there was amplification in the Bwr-6 QTL. Contrary to the morphological studies, two accessions (GC and GG) classified as resistant were identified as highly susceptible to the pathogen under field conditions. The difference in morphological and molecular categorization as obtained in this study is consistent with Olasanmi et al.[28] who reported inconsistencies in cassava mosaic disease resistance levels in cassava genotypes based on field and molecular marker data. Accessions classified as resistant based on molecular markers but identified as susceptible under phenotypic evaluation according to Wang & Lin[29] may be attributed to the presence and interaction of other strains of the R. solanacearum on the field, inoculum density, soil moisture, temperature, and presence of root-knot nematodes which can affect the stability of bacterial wilt resistance in crop genotypes.
-
The results presented in this study show variations in the reaction of different accessions to bacterial wilt disease using both phenotypic and molecular markers. Using only phenotypic scores two accessions were classified as moderately resistant while three accessions were selected as resistant based on the molecular markers used. However, only accession L_020 phenotypically identified as moderately resistant was further confirmed as resistant based on the molecular data and therefore, selected as a promising genotype to be exploited as a potential rootstock to manage bacterial wilt disease. Due to the inconsistent phenotypic and molecular data obtained in this study, accessions (CRI-04, GG, and GC) cannot be selected as potential rootstocks to manage bacterial wilt disease in tomato production. These accessions can be screened further with additional molecular markers and under different fields to confirm the results obtained.
-
The authors confirm contribution to the paper as follows: performing the research: Adomako J, Prempeh RNA; data analysis and technical help: Osei MK, Gyau J, draft of the manuscript: Cho MC, Adomako J, Prempeh RNA, Osei MK; experiments design, study supervision and manuscript revision: Boakye-Mensah IN, Osei-Bonsu I, Ofori P. All authors reviewed the results and approved the final version of the manuscript.
-
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
This study was supported by a grant from KAFACI, Rural Development Administration of Korea (Grant No. KAH2000106).
-
The authors declare that they have no conflict of interest.
- Copyright: © 2024 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Adomako J, Osei MK, Prempeh RNA, Osei-Bonsu I, Gyau J, et al. 2024. Identification of Ralstonia solanacearum resistant solanum plants as potential rootstock to manage bacterial wilt disease in tomato production. Technology in Horticulture 4: e020 doi: 10.48130/tihort-0024-0017
Identification of Ralstonia solanacearum resistant solanum plants as potential rootstock to manage bacterial wilt disease in tomato production
- Received: 13 May 2024
- Revised: 25 July 2024
- Accepted: 29 July 2024
- Published online: 04 September 2024
Abstract: Using morphological and molecular markers, this study screened tomato (Solanum lycopersicum) and garden egg (Solanum melongena) accessions for resistance to bacterial wilt disease. The solanum plants were inoculated with Ralstonia solanacearum and evaluated for disease incidence and severity in a field trial set up in a Randomised Complete Block Design with four replications. Molecular markers conferring resistance to R. solanacearum Phylotype I and II were used to identify durable and partial resistance. Results showed significant variation in disease incidence and severity among accessions, with tomato accessions exhibiting higher susceptibility. Accession CRI-01 had the highest disease incidence (54.0%), while accession GD had the lowest (13.0%). Accession CRI-04 showed moderate resistance with a disease severity index of 0.37, while accession GC had the highest disease severity index (0.90). Accession L_020 demonstrated moderate resistance in the field (43.0% disease incidence) and possessed durable resistant genes, making it a promising rootstock for managing bacterial wilt disease in tomato production. This research contributes to the development of integrated pest management strategies for sustainable tomato production.
-
Key words:
- Bacterial wilt /
- Garden eggs /
- Resistant rootstock /
- Integrated pest management /
- Tomato /
- Sustainable production












