-
Deep learning, a subfield of machine learning, is distinguished by its computational model's capacity to acquire knowledge from abstract data using structures consisting of multiple processing units[1]. Such models use automatic optimisation of model parameters, e.g. stochastic gradient descent, batch gradient descent, Adam optimiser, to systematically optimise the basic parameters of the data computation within the model architecture to achieve the goal of optimising or accelerating the optimization of the model parameters. The automatic optimisation of model parameters eliminates the need for manual design of parameters within the model and reduces the amount of manual work involved in model development. In addition, the trained models can be used to achieve objectives such as object detection, localization, image classification, and predictive analytics based on complex abstract data. One noteworthy advantage of contemporary machine learning methodologies is their inherent capability for automated feature extraction and learning from abstract data, thereby obviating the requirement for manual intervention in guiding the model through the processes of feature extraction and learning, as demonstrated in conventional feature engineering. In contrast, traditional machine-learning approaches necessitate the construction of relevant features by humans, contingent upon the dataset, a process commonly referred to as feature engineering. Performing the task of feature engineering is inherently complex and time-consuming, necessitating iterative human adjustments based on changes in the dataset or design requirements[2]. Traditional feature engineering may be more advantageous when dealing with small datasets is required. But in today's environment of increasing labour costs[3] and decreasing computer arithmetic costs[4], as well as in practice where large neural networks based on deep learning can be better generalized this is more important[5]. Deep learning methods may be more advantageous when dealing with large datasets or when a high degree of automation is required. As a result, the developmental costs associated with models of this kind, which is mainly labour costs, significantly increase, while their generalizability is concurrently limited.
In contrast, the incorporation and enhancement of network models which involve proficient feature extraction techniques combined with deep learning, are exemplified by Convolutional Neural Networks (CNN), You Only Look Once series (YOLO), Single Shot Multibox Detector (SSD), Residual Network (RstNet), Densely Connected Convolutional Network (DenseNet), GoogleNet, MobileNet, and Xception, have significantly enhanced the automatic feature-learning capabilities of models. These advancements enable deep-learning models to autonomously extract features of varying levels of complexity from raw data, displaying significant potential for improving the reliability of models based on leaf disease image analysis. As a result, models developed within the deep learning framework align with the processing of intricate abstract data, such as images, and it also demonstrates advantages in terms of labour costs, expenditure on productivity costs, and increased model versatility[6].
Early detection of diseases minimizes the overuse of pesticides in disease prevention[7]. The utilization of deep learning techniques for monitoring crop leaf diseases enable the analysis of various disease types by inputting images of diseased leaves into a model previously trained for these leaf diseases, eliminating the necessity for specialized personnel and allowing for an automated identification process, that enables rapid, timely control[8]. When these trained models are deployed on small mobile terminals, it helps non-professional agriculturists to detect problems in time, even in completely unmanaged farmland, so that preventive measures can be implemented[9]. Due to the good generalisation ability of the model, identification aided by the use of a trained model can provide a more reliable identification reference for experts[10]. For example, in changing environments where some leaf diseases have no obvious symptoms or are difficult to detect, which requires plant pathologists to be well observed[11], deep learning models can extract features that are difficult to observe with the human eye[12]. Thereby, the application of such technologies contributes to the timely detection and prevention of plant leaf diseases, optimization of crop yields, and advancement of precision agriculture. Consequently, this enhances productivity, reduces labour costs, and strengthens sustainability[13].
Tropical areas hold a significant position as major producers of staple food crops such as rice, corn, and millet, as well as various fruit crops including banana, mango, coconut, and durian. The region's consistently high temperatures lead to shorter pathogen incubation periods, exemplified by the reduced diurnal temperature which accelerates the latency period of pathogens like Hemileia vastatrix, causing rust epidemics in Central America[14] Furthermore, the typically high humidity in tropical regions foster both crop growth and the survival of harmful bacteria[15]. Consequently, tropical regions experience more frequent occurrences of plant diseases, posing a threat to food security[16]. Thus, compared to non-tropical regions, crops in tropical areas have shorter growth cycles and are more susceptible to rapid disease outbreaks, underscoring the necessity for timely and effective plant disease prevention and control measures[16]. Because deep learning-based leaf disease detection technology can be mounted on mobile devices and achieve real-time monitoring, this technology is more suitable for tropical areas.
The current study focuses on the application of detection technologies for plant leaf diseases and pests, exploring contemporary technological methodologies and attributes underpinning current crop leaf disease and pest detection techniques. This paper describes the technical characteristics and advantages of parallel detection using deep learning models. The development direction and challenges of deploying deep learning-based leaf disease detection technology in tropical regions have also been discussed.
-
Leaf disease detection can be regarded as the detection of leaf disease objects, which is considered as a computer vision task, involving two key targets: object recognition (classification) and object localization. The process of building object detection can be divided into three main parts: data set selection or construction, model selection and training, model evaluation and deployment.
Choosing the right architecture for the detection model depends primarily on the required level of accuracy, inference speed, and available computational resources. The detection methods are distinguished by whether object localization and classification are performed in a single stage[17]. The one-stage methods directly predict bounding boxes and class probabilities for all objects in a single pass without region proposal[18], such as YOLO series, SSD, CornetNet, et al. However, the tow-stage methods are much more complicated, like CNN, RCNN. The first stage usually proposes potential regions of interest, and the second stage refines these proposals by classifying and adjusting the bounding boxes. This stage difference in practical application leads to the two models having different emphasis on prediction speed and prediction accuracy[19], which the one-stage method emphasizes the prediction speed, correspondingly, and the other emphasizes the prediction accuracy. Popular CNN architecture used as the backbone includes AlexNet and its variants like VGGNet, GoogLeNet, Inception series, ResNet and its variants like ResNet50 and ResNet101, DenseNet, MobileNet.
Currently, based on the literature summarized in Tables 1 & 2, the authors' models have consistently achieved recognition accuracy exceeding 90% in validation or prediction applications. A 90% recognition accuracy in target recognition models is often seen as high, but its adequacy depends on the specific application and requirements. Thus, there isn't a universal benchmark, and performance evaluation should align with specific tasks and applications[20]. Some models even exhibit recognition accuracy surpassing 99% in test sets. In the realm of laboratory development and certain outdoor applications, achieving high accuracy with simple models is still a prominent challenge[21]. Furthermore, due to the diverse hardware platforms employed by different authors and the specific purposes of their applications, direct comparisons of reasoning speed and computing resource requirements among various models pose challenges. Nevertheless, a delicate balance is required between recognition accuracy, model size, and model inference speed. This becomes particularly critical when contemplating the deployment of models on mobile terminals for practical applications. Achieving this balance represents a formidable challenge in the field.
Table 1. Plant disease open data sets.
Data set name Crop Brief description Ref. Image Database of Plant Disease Symptoms 21 plant species In October of 2016, this database called PDDB, had 2,326 images of 171 diseases and other disorders affecting 21 plant species including soybean, citrus, coconut tree, dry bean, cassava, passion fruit, corn, coffee, cashew tree, grapevine, oil palm, wheat, sugarcane, cotton, black pepper, cabbage, melon, rice, pineapple, papaya, cupuacu. [26] Tomato leaf disease detection Tomato A tomato leaf disease dataset includes tomato mosaic virus, tomato yellow leaf curl virus, late blight, leaf mold, early blight, septoria leaf spot, Spider mites Two-spotted spider mite, healthy tomato. [27] Agricultural Disease Image Database for Agricultural Diseases and Pests Research Rice and wheat, fruits
and vegetables, etc.The dataset currently has about 15,000 high-quality agricultural disease images, including field crops such as rice and wheat, fruits and vegetables such as cucumber and grape, etc. [28] Rice disease dataset Rice A rice leaf disease dataset includes bacterial leaf blight, blast and brown spot. [29] Pathological images of apple leaves Apple The apple leaf disease image dataset contains 8 common apple leaf diseases: Mosaic, rust, gray spot, mottle leaf disease, brown spot, black star disease, black rot, healthy leaf disease. [30] Plant Pathology 2021 - FGVC8 Apple 2021-FGVC8 contains approximately 23,000 high-quality RGB images of apple foliar diseases, including a large expert-annotated disease dataset. [31] Tomato Disease Multiple Sources Tomato Over 20 k images of tomato leaves with 10 diseases and one healthy class. Images are collected from both lab scenes and in-the-wild scenes, which includes late blight, healthy, early blight, septoria leaf spot, tomato yellow leaf curl virus, bacterial spot, target spot, tomato mosaic virus, leaf mold, spider mites Two-spotted spider mite, powdery mildew. [32] Data for: Identification of Plant Leaf Diseases Using a 9-layer Deep Convolutional Neural Network 12 plant species The dataset includes 39 different classes of plant leaf and background images are available. The data-set contain 61,486 images. [33] Table 2. Leaf disease detection technology and corresponding advantages.
No. Model name Technical characteristics Advantages. Ref. 1 Apple-Net The Feature Enhancement Module (FEM) and Coordinate Attention (CA) incorporation.
Generative Adversarial Networks (GAN) for interference reduction.Multi-scale information acquisition, increased diversity, and noise resistance. [34] 2 Mobile Ghost with Attention YOLO Ghost modules and separable convolution for reducing model size.
The mobile inverted residual bottleneck convolution with Convolutional Block Attention Module (CBAM) for improving feature extraction capability.Lightweight real-time monitoring (10.34 MB), suitable for mobile terminals. [35] 3 BTC-YOLOv5s Bidirectional Feature Pyramid Network (BiFPN) for a fusion of multi-scale features.
Transformer attention mechanism for capturing global contextual information and establishing long-range dependencies.
CBAM for interference reduction.Reduces irrelevant information, small model size (15.8 MB). [36] 4 AlAD-YOLO The backbone network of TOLOv5s replaced with that of MobileNetV3. Reduction in parameters and computational complexity during feature extraction. [37] 5 YOLOX-ASSANano Asymmetric ShuffleBlock for enhanceing feature fusion.
Cross stage partial module with shuffle attention for interference reduction.Processes complex natural backgrounds and lightweight model. [38] 6 V-space-based Multi-scale Feature-fusion SSD Multi-scale attention extremum for automatic lesion detection. Enhances detection ability for disease lesions, especially small ones. [39] 7 LAD-Net Asymmetric and dilated convolution as the convolution to reduce model size.
LAD-Inception designed with an attention mechanism for improving multiscale detection capabilities.Small model size (1.25 MB), high accuracy (97.72%), and implementation of deployment on mobile devices. [40] 8 Enhanced LSTM-CNN Majority voting ensemble classifier replaced the classifier.
Optimal LSTM layer network applied to select deep features autonomously.Enhanced feature extraction and classifier modification. [41] 9 LALNet EARD module with multi-branch structure and depth separable modules extracts more feature information with fewer parameters and computational complexity. SE attention module for increase the feature extraction capability. Small size (6.61 MB), fast execution (6.68 ms/photo), and high recognition accuracy. [42] 10 Two-stage detection system Three-way classification in the first stage using Xception as the base model.
Real-time detection in the second stage.Detects multiple diseases with 87.9% mean average precision. [43] 11 Improved Faster R-CNN Res2Net and feature pyramid network replaced the backbone of Faster R-CNN for batter feature fusion.
RoIAlign instead of RoIPool of Faster R-CNN for improving the identification precision.Extracts multi-dimensional features in natural scenes with complex backgrounds. [44] 12 BC-YOLOv5 Modify YOLOv5 neck structure with weighted BiFPN and CBAM. Enhanced feature extraction in the detection layer, reduced irrelevant information for complex backgrounds. [45] 13 PLPNet Perceptual adaptive convolution (PAC) for enhancing the
network's global sensing capability.
location relation attention module (LRAM) for reducing unnecessary information.
SD-PFAN structure for fusing features batter.Recognizes leaf diseases at the edge of the leaf, resist background interference. [46] 14 DL Technique U-net with Gradient GSO for leaf segmentation in the first stage.
DbneAlexnet trained using proposed GJ-GSO for leaf classification using Gradient Jaya-Golden search optimization in the second stage.Two-stage approach mitigating background noise. Optimized segmentation and classification through new training methods. [47] 15 LightMixer Depth convolution with Phish (DCWP) and light residual (LR) modules to increase feature integration and reduce parameters.
Phish activation function for reducing the information loss.Identifies diseases in complex environments, suitable for mobile deployment. [48] 16 NanoSegmenter Transformer structure and sparse attention mechanisms to tackle the instance segmentation task, replacing the CNN backbone.
The bottleneck inversion technique to achieve model lightweighting.High accuracy in instance segmentation, low computational complexity, and small model size. [49] 17 DMCNN Multi-scale convolution for disease classification from multiple channels. Enhancement of accuracy and efficiency through multi-scale detection [50] 18 CRNN Combines CNN and RNN for improved sequential features extraction. Achieves significant improvement in maximum accuracy compared to traditional CNN. [51] 19 Transfer learning with pre-trained CNN models. Transfer learning with Faster-RCNN and Inception ResNetv2 models. High recognition ability on new dataset after transfer learning. [52] 20 PCA DeepNet Data enhancement with CycleGAN
Feature extraction with PCA
Classification with Faster-RCNN.Innovative PCA method for image extraction, followed by Faster-RCNN for classification. [53] 21 Four transformer-based models. Comparative study on four vision transformers (EANet, MaxVit, CCT, PVT) for tomato leaf disease identification. MaxViT architecture identified as the best for tomato leaf disease identification. [8] 22 Fine-grained image identification framework Utilizes OPM, DRM, AADM, and OCB for object identification, feature learning, and severity assessment. Assess severity based on categorized dataset, captures fine-grained details with DRM. [54] 23 RiceNet YOLOX identifies disease sites in the first stage.
Siamese Network classifies diseases in the second stage.Effective two-stage detection, addressing complex backgrounds and limited samples. [55] 24 RWW-NN SetNet isolates the rice crop images.
RWW algorithm (WWO & ROA), for improved classification.Two-stage approach mitigating background noise, improved classifier performance. [56] 25 The domain adaptation networks with novel attention mechanisms Channel and spatial attention mechanism (CPAM) in DSAN for key feature identification. Alleviates data distribution differences and small sample problems. [57] 26 RiceDRA-Net Res-Attention module based on CBAM for accurate disease identification and localization.
DenseNet-121 serves as the backbone network.Precise disease localization, even in complex backgrounds. [58] 27 rE-GoogLeNet ECA attention mechanism in GoogLeNe
Residual networks for information loss mitigation.Improved recognition and performance over alternatives. [59] 28 ADSNN-BO Enhanced self-attention mechanism employed along the entire architecture in MobileNetV1,
Bayesian optimization for hyperparameter tuning.Outperforms MobileNet with 3.6% accuracy improvement. [60] 29 DGLNet Global attention module (GAM) enhances sensitivity by reducing background noise.
Dynamic representation module (DRM) for flexible feature acquisition.Enhances generalization capability and feature representation in lightweight models. [61] 30 Novel rice grade model EfficientNet-B0 architecture as the backbone for better recognition accuracy for spotting diseases.
By identifying leaf instances and disease areas, the ratio of the two areas was calculated to estimate the severity of the disease.Reliable disease spot recognition, quantifies severity of rice disease. [62] 31 Comparison of pre-trained residual network models Comparison of ResNet34, ResNet50, ResNet18 with self-attention and ResNet34 with self-attention. Models with self-attention exhibit improved recognition accuracy during transfer learning. [63] The model's ability to recognize is strongly tied to the quality and specifics of the dataset used for training, validation, and testing. For instance, factors such as the accuracy of object labels, the number of images and labels in the data set, balanced data distribution, and the use of data enhancement methods impact the ultimate prediction accuracy and the model's ability to generalize. He et al.[22] suggested that imbalanced data could compromise model classification accuracy. The size of the dataset is commonly regarded as significant which is the number of images and labels in the data set, yet it is not definitive[23]. While expanding the dataset size may enhance the model's generalization capability and performance to some degree, it does not imply that a larger dataset always leads to better outcomes or more accurate identification under identical conditions[24]. Errors in data labels can significantly impact the accuracy of model testing. These views are further extended and proposed by Priestley et al.[25] and they pointed out differences in data management practices between academia and industry, underlining that data quality should align with diverse user needs. They also proposed that the availability of data can be supported by an infrastructure for data collection and management, particularly in large organizations. In academia, data sets are typically categorized as published or non-public, posing challenges in objectively assessing the quality of datasets used in some researcher-proposed models.
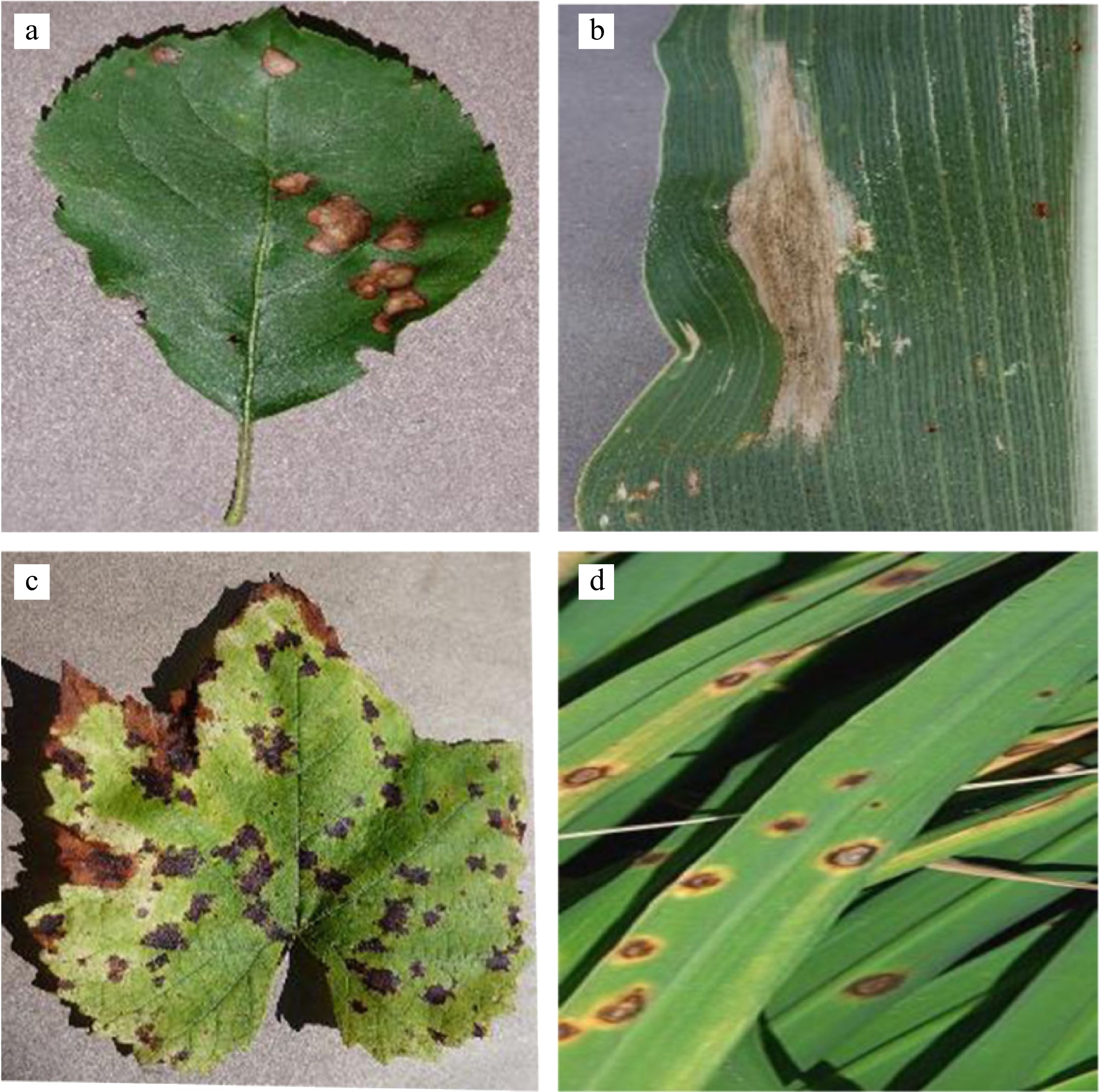
In practice, an important consideration influencing model performance is whether images in the dataset capture natural environments, encompassing both indoor and outdoor scenes. For example, as shown in Fig. 1a−c, are all indoor, d is outdoor. Natural environment photos tend to have complex backgrounds, demanding models with stronger anti-interference capabilities against intricate scenarios. As researchers develop models, it is still suggested that if the researchers want to develop a model for the identification of a leaf disease, it is a very necessary process to collect relevant data and label it. The research focus of tropical leaf disease detection is still based on effective data collection. Based on datasets from Tables 1 & 2 this article organizes the published datasets, including referenced ones, for further reference.
-
A review was conducted on a corpus of scholarly papers abput leaf disease detection published in the year 2023. The focus of the review includes the characteristics and advantages of techniques for detecting leaf disease using deep-learning models. The selected papers collectively encapsulated a broad spectrum of contemporary deep-learning models employed for the detection of leaf diseases. This article provided detailed insights into the characteristics of these models. Additionally, the review meticulously cataloged the salient features utilized of the model, thereby affording a understanding of the state-of-the-art methodologies employed in the domain of leaf disease detection. The models used in this study include models based on mature algorithm technology models such as YOLO, SSD, and CNN. The investigator has undertaken pertinent modifications to these infrastructures, tailoring them to optimize their efficacy for the distinct application conditions posed by the identification of pathological manifestations in foliage, including methods of identifying leaf disease using first and second-stage models.
These models exemplify superior accuracy, lightweight architecture, and adept deployment on mobile devices, rendering them well-suited for the detection of leaf diseases across diverse scenarios. Notably, enhancements in recognition accuracy are achieved through the replacement of backbone networks or the introduction of innovative modules. These improvements are usually to enhance the ability to extract features from images or the ability to fuse features after extraction, and to reduce interference as elucidated in Table 2 (No. 1, 2, 3, 5, 6, 9, 11, 12, 13, 16, 17, 18, 20, 26, 27, 30). Additionally, attention mechanisms play a pivotal role in refining recognition accuracy by focusing on key information on different channels or convolution kernels at different scales, as evidenced by the methodologies delineated in Table 2 (No. 1, 2, 3, 5, 6, 7, 9, 12, 13, 16, 17, 18, 22, 25, 26, 27, 28, 29, 31). Furthermore, after comparing the four state-of-the-art(SOTA) Vision Transformer models by Hossain et al.[8], it is concluded that MaxViT has better recognition accuracy, which proved that using global attention is more suitable to improve the recognition accuracy of the leaf disease identification.
Techniques for refining input image quality, exemplified by the implementation of Generative Adversarial Networks (GAN)[34,53], further contribute to the augmentation of accuracy. Finally, introducing other methods to improve the classifier can also increase the accuracy of recognition[41,56] and optimize the training method of the model[47].
Moreover, as part of the overarching goal to enhance recognition accuracy, the adoption of pre-trained models through transfer learning on newly curated datasets emerges as a commendable paradigm, as advocated by Saeed et al.[52], Simhadri & Kondaveeti[64] and Sudhesh et al.[63]. The imperative for reduction in model size is achieved through the introduction of novel modules or the replacement of the feature extraction network, as evidenced by instances in Table 2 (No. 2, 4, 7, 9, 15, 16).
Furthermore, certain authors have made notable contributions to practical field applications, exemplified by endeavors in real-time processing, the amelioration of environmental challenges and assessment of disease severity. The two-stage methodology includes classification before detection and detection before classification, demarcating the detection process into initial leaf segmentation and subsequent leaf disease classification, as undertaken by Khan et al.[43] and Badiger & Mathew [47], exhibits a proclivity towards effectively discerning multiple diseases on leaves. Pen et al.[55] and Daniya & Vigneshwari[56] also used a two-stage approach to solve the problem of disease identification from photos with complex background obtained in actual fields, which can overcome the interference of complex background environments to recognition to some extent. At the same time, because a new segmented disease data set is generated in the process, the problem of small samples with fewer original images is also solved.
Leaf diseases of tropical plants usually have the following characteristics: a variety of leaf diseases, rapid outbreak of leaf diseases, high frequency, and difficult to prevent. In addition, unlike in other regions, tropical plants tend to be relatively tall, with thicker foliage and a faster growth cycle. This makes disease surveillance and management of tropical plants more difficult. For some fruit crops, such as coconut, mango, lychee, and durian, it is difficult to achieve early detection and early treatment of leaf disease. Even though some crops can be reduced in height through dwarfing management, they still have wider leaves for relatively similar crops such as apples and tomatoes. This makes it difficult to use a camera to photograph the diseased leaf in its entirety up close. Therefore, new requirements are put forward for leaf disease detection technology based on deep learning models especially for real-time monitoring.
Given the difficulties in the detection of leaf diseases of tropical plants like coconut, such as the small number of leaf disease data sets, the mutual occlusion of large leaves, the influence of leaf shadows, and the interference of complex leaf backgrounds, some researchers have conducted studies. Thite et al. have published a dataset named 'Coconut (Cocos nucifera) Tree Disease Dataset', which contains five diseases: Bud Root Dropping, Bud Rot, Gray Leaf Spot, Leaf Rot, and Stem Bleeding[65]. The images in this dataset are centered on disease locations and also include disease photos presented on tree trunks. In addition, researchers have developed a detection model for coconut tree disease. The model uses the newly developed AIE-CTDDC technology[66]. To solve the problem of identifying coconut tree disease in the complex coconut leaf background environment, the model uses CapsNet[67] as the feature extractor, and the data is pre-processed using MF-based enrollment removal technology before this. Similarly, to solve the problem of mutual occlusion of large coconut leaves and the impact of leaf shadows on the recognition effect, Subbaian et al. proposed a coconut leaf disease detection method based on YOLOv4[68]. The method improved the prediction accuracy of the model through multi-scale detection, PANet, and adaptive border improvement.
In terms of the portability of detection and solving the problem that the plants are too high to observe, some researchers have proposed some methods and applications for the detection of durian leaf disease. Gallenero & Villaverde designed a portable device embedded with the Duri Premium application to identify durian leaf disease. The device was equipped with the Mobilenet-based convolutional network model, which achieved good identification accuracy[69]. Also for portable detection, a mobile application was developed to detect the leaf diseases of mango and grape by Rao et al.[70]. To solve the problem caused by the rapid detection and prevention of durian leaf disease, Piriyasupakij & Prastiphan designed an unmanned aircraft equipped with YOLOv5 for the detection of durian leaves on the tree and realized the effect of automatic cruise shooting and identification of durian leaf disease[71]
From the above, it can be concluded that when identifying leaf diseases in tropical crops, researchers need to consider two aspects. On the one hand, it is to solve the impact of large blade occlusion and complex leaf surface environment around leaf disease. Another aspect is that to achieve rapid leaf disease detection, it is necessary to carry out portable design of detection equipment, such as mounted on mobile terminals, to cope with complex detection environment or to detect excessively high plant leaf disease.
-
There is a trade-off between the speed and accuracy of model inference. In general, increasing the inference speed of a model may result in decreasing the recognition accuracy of the model, and vice versa. This is because when designing a model, to improve the reasoning speed, it is often necessary to reduce the complexity and the number of parameters of the model, which may lose certain recognition accuracy. On the contrary, to improve the recognition accuracy of the model, it may be necessary to increase the complexity and the number of parameters of the model, resulting in slower inference speed.
To assess and compare the performance of the models, the model prediction results are commonly used as True Positive (TP), which refers to the number of positive samples correctly identified; False Positive (FP), which refers to the number of negative samples incorrectly identified; True Negative (TN), which refers to the number of negative samples correctly identified; and False Negative (False Negative, FP) refers to the number of negative samples that are incorrectly identified. The correspondence is shown in Table 3.
Table 3. Classification of predicted and actual results.
Actual Predicted Positive Negative True True positive True negative False False positive False negative Based on the four results of sample classification, it also sets Accuracy to indicate the samples predicted to be classified correctly among all samples; sets Precision to indicate the proportion of true-positive samples among those predicted to be positive; sets Recall to indicate the proportion of true-positive samples among those classified correctly; and sets the average precision. The corresponding formulas for Accuracy, Precision and Recall are as follows, respectively.
$ Accuracy=\dfrac{T P}{T P+F P+T N+F P} $ (1) $ Precision=\dfrac{T P}{T P+F P}$ (2) $ Recall=\dfrac{T P}{T P+F N}$ (3) In the assessment of the model classification quality of each category, since the use of quasi-departure rate, checking rate, and recall rate alone cannot be considered together to assess the score, the researcher, therefore, proposes the use of Average Precision (AP) as a measure of the quality of the model classification for a certain category, i.e., integrating a function plotted on a certain category of objects with the Recall (r) as the horizontal axis and the corresponding Precision (p(r)) as a function plotted on the vertical axis for integration; use of mean Average Precision (mAP) for evaluating a model for multiple classes (n) of object classification performance evaluation metrics; F1 Score is used as an assessment of the combined consideration of check accuracy and recall, i.e., the reconciled average of check accuracy and recall. The Average Precision and F1 Score correspond to the following calculation formula:
$ AP=\underset{0}{\overset{1}{\int }}p\left(r\right)dr $ (4) $mAP=\dfrac{1}{n}{\sum }_{i=1}^{n}A{P}_{i} $ (5) $ {F}_{1}=\dfrac{2Precision\;\times\; Recall}{Precision\;+\; Recall}$ (6) In improving the accuracy of recognition, there are limitations in the identification of leaf disease using two-dimensional image processing. The approaches mentioned in Table 2, which employ convolutional and deep learning networks for image feature extraction, are inherently designed for the analysis of two-dimensional images. In practical applications, foliage afflicted with diseases often exhibits characteristics such as leaf curl, damage, and instances of mutual occlusion during the acquisition of field imagery. Consequently, a nuanced examination of disease severity, based on a model learned on two-dimensional image data alone, predicated on estimating the proportion of the diseased area relative to the entire leaf surface[62], may lead to the inadvertent[72]. Researchers have suggested a method for creating three-dimensional reconstructions using two-dimensional images, aiming to overcome spatial limitations present in these types of pictures. The research shows that it is feasible to deduce crop height and leaf area through 3D modeling[73]. Compared with the 2D RGB image processing method, the 3D method can accurately estimate the number of leaves, avoid the influence of mutual occlusion of leaves to a certain extent, and greatly improves the accuracy of detection[74]. At the same time, it may also solve the problem proposed by Tang et al.[46], that the occurrence of diseases at the edge of leaves in a complex background will interfere with the recognition. Utilizing the approach of reconstructing a three-dimensional model based on two-dimensional images still poses challenges[75−77]. These challenges encompass the loss of depth information, compromised accuracy due to low resolution or distorted images, and difficulties in precisely capturing intricate geometric textures, particularly in complex scenes. In terms of computing cost, it cannot be ignored that three-dimensional method consumes more computing cost than two-dimensional method[78].
In addition, to improve the quality of recognition, the solution of the multi-scale detection problems and the application of the attention mechanism have played a great help. Objects of all sizes (objects proportional to the size of the image) need to be detected, requiring the network to have the ability to recognize objects of different sizes, faced with the challenge of significantly decreasing detection accuracy for very large or very small scale targets[36]. However, the deeper the network, the smaller the size of the feature map, which makes it difficult to detect small objects, which is a problem that cannot be avoided after the model extracts the feature map[79]. This problem can be alleviated in the process of extracting features[39] and in the process of feature fusion[80], to improve the average precision of the model. The attention mechanism is a self-supervised learning method used in the natural language processing, and applied to enable the network to focus on the target region with important information by learning how much the input data contributes to the output data, while suppressing other irrelevant information and reducing the interference caused by irrelevant background on detection results[36,81]. This method can be applied to the model to extract features of different channels, for example, CBAM[82], BAM[83].
The recognition speed of the evaluation target recognition model usually has the following evaluation indexes, such as inference time, inference throughput, inference frame rate, and hardware resource utilization. These evaluation indexes are also used to evaluate the reasoning speed of the model in specific application scenarios. Inference time is commonly used to evaluate the speed of the model in image recognition and classification, which means the time it takes the model to go from receiving the input image to outputting the prediction.
In speeding up the prediction speed, the one-stage target detection method has more advantages. For the one-stage recognition method based on YOLO, the anchor method should be used for frame selection first, especially for YOLOv2 to YOLOv6, which takes up computing resources. To reduce model size and prediction speed, many researchers proposed an anchor-free method, which takes key points as the core. For example, the target center of the feature map is taken as the key point to locate the target. Based on the number of key points, the free-anchor method can be divided into central-point-based method and multi-key point-based method, such as CenterNet[84]. From a hardware aspect, different hardware selected for the prediction means different predictive speeds under the same model selection and parameters[85].
In practical production, a single plant can exhibit concurrent occurrence of multiple diseases, with distinct characteristics of various diseases observed on the same leaf, or the same disease has different characteristics at different times[86]. This multifaceted infection pattern can be considered a more indicative measure for assessing current crop damage levels, placing higher demands on the accuracy and generalization capacity of disease identification models.
Overall, if the leaf disease identification method is deployed in practical production, the balance between recognition accuracy and recognition speed can not be achieved only by optimizing the model or hardware. However, in terms of the evolution and development of methods for identifying leaf diseases using convolutional neural networks, the balance between accuracy and speed has to be mentioned.
There are still some ways to balance the relationship between speed and accuracy of model inference. These methods can be roughly divided into three general directions: model compression, hardware optimization, and algorithm improvement. Model compression[87], such as channel pruning[88] and knowledge distilling[89], can reduce the number of parameters and complexity of the model, thereby improving the model reasoning speed and maintaining the accuracy of recognition to a certain extent[87]. Hardware optimization can often significantly increase the speed of model inference. For example, running a model on a GPU, TPU, or professional computing device can significantly increase the speed of the model inference[90]. There are more methods to accelerate the model inference speed and improve the inference accuracy by improving the algorithm, which are not listed in this paper. All three methods can improve the performance of the model both during training and during inference.
It is noteworthy that the lower the error rate of the training model is not equal to the better the quality of the model when training models and too low a classification error rate usually leads to overfitting problems. For example, such as a fully connected network classifier, one should not simply assume that achieving the best learning quality is synonymous with minimizing the classification error rate. Some researchers have delved into understanding the delicate balance between learning difficulty and learning speed. By utilizing a single-layer perceptron and a double-layer neural network optimized with a gradient descent learning algorithm, the average accuracy typically decreases with training time. The model attains a harmonious equilibrium between training difficulty and learning rate when the training error rate is at 15.87%[91], resulting in an approximately 85% accuracy.
Hence, in the pursuit of a specific characteristic index for the model, it is imperative to selectively adjust and optimize the model based on the prevailing circumstances or specific requirements. Furthermore, relying on a singular index is inadequate for evaluating the overall quality of a given model.
-
In the realm of agriculture, the identification of crop diseases stands as a pivotal task, serving as a key to further assessing the severity of current or potential hazards. The foregoing review elucidates that the application of artificial intelligence (AI) technology in monitoring plant leaf diseases attains commendable levels of recognition accuracy and expeditious identification in the model development and testing. This methodology emerges as a proactive approach to disease identification, conferring the capacity to empower agricultural stakeholders and experts in effectually addressing extant diseases or preemptively mitigating potential threats. Moreover, through the implementation of smart agriculture methodologies, the attainment of sustainable and resilient production is conceivable, thereby mitigating environmental impact and fortifying food security[92]. The judicious quantification of crop diseases fosters the formulation of precise protection strategies tailored to the dynamic and perpetually changing agricultural milieu. This approach facilitates the adoption of targeted disease prevention and control measures, consequently diminishing the superfluous use of pesticides. The resultant abatement in pesticide application not only serves to curtail production costs but also mitigates environmental pollution arising from pesticide usage[93]. The application of artificial intelligence (AI) technology for identifying leaf diseases, while promising, is not without potential challenges.
In the application of tropical plant leaf disease recognition, the real-time monitoring and mobile device support characteristics based on deep learning model can greatly solve the characteristics of tropical plant leaf disease difficult to find and observe in time. To further solve the problem of insufficient computing power of mobile hardware devices or high demand for model recognition accuracy, the Master-Slave structure can be used. In this structure, the master model acts as the central node, such as the cloud platform, responsible for coordinating and controlling the operation of the whole system, while the receiver acts as the slave node, such as mobile devices, responsible for receiving and processing the instructions or data of the autonomous model, which can effectively realize the parallel processing and collaboration of tasks.
Current studies have shown that terahertz waves can be used to detect physiological and biochemical parameters in plant leaves, such as water content[94], chlorophyll content, cell structure, and cell wall thickness, to indirectly reflect the occurrence and development of leaf diseases. Terahertz waves are electromagnetic waves between microwaves and infrared light, with frequencies ranging from 300 GHz to 3 THz. Terahertz waves have a wide range of applications in biomedicine, material science, and safety testing. Terahertz waves have strong penetration in biological materials and are also resonance absorbed by biomolecules, so they can be used to detect changes in the internal structure of plant leaves and biomolecules. It is possible to use deep learning models to analyze the signals of crop leaves fed back by terahertz waves, but the technology is still in the research stage[95,96]
The rapid development of large language modeling in recent years has made it possible to combine large language modeling with leaf disease detection techniques[97]. The rapid development of large language modeling in recent years has made it possible to combine large language modeling with leaf disease detection techniques. Large language models have excellent advantages in processing and analyzing literature and data, which can help researchers better understand and grasp the research progress within the field of leaf disease detection. In addition, the powerful text comprehension and text generation capabilities of the big language model can assist in the annotation and enhancement of plant petiole image data, thus improving the efficiency of model training. Similarly, the Big Language Model can assist in reasoning and summarizing the results of leaf disease detection and provide scientific references and bases.
However, it is still due to the complex and changeable climate environment such as tropical high temperatures and high humidity, especially considering the production and investment costs, it is not practical to use mobile devices equipped with identification models for detection. Hot and humid environments tend to damage electronic equipment, which increases maintenance costs after the equipment is deployed to the field.
-
The application of artificial intelligence (AI) technology in the detection and diagnosis of crop leaf diseases represents an advanced approach in precision agriculture, which particularly in machine learning and deep learning, various methods have proven effective in automating the identification and classification of crop leaf diseases. However, in practical implementation, it is imperative to carefully choose the suitable model and method for deployment based on the specific circumstances and demands. The detection and management of plant diseases in tropical areas remain a multifaceted issue. This technological application aims to swiftly and accurately evaluate the situation, thereby enabling timely interventions to mitigate the adverse impact of diseases on crop productivity.
-
The authors confirm contribution to the paper as follows: study conception and design: Huang M; manuscript preparation: Yao Z, Huang M. Both authors read and approved the final version of manuscript.
-
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
This research was supported by the Major Science and Technology Project of Hainan Province (Grant No. ZDKJ202017-2-2).
-
The authors declare that they have no conflict of interest.
-
Received 6 January 2024; Accepted 15 April 2024; Published online 26 June 2024
- Copyright: © 2024 by the author(s). Published by Maximum Academic Press on behalf of Hainan University. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Yao Z, Huang M. 2024. Deep learning in tropical leaf disease detection: advantages and applications. Tropical Plants 3: e020 doi: 10.48130/tp-0024-0018
Deep learning in tropical leaf disease detection: advantages and applications
- Received: 06 January 2024
- Revised: 07 April 2024
- Accepted: 15 April 2024
- Published online: 26 June 2024
Abstract: This paper delves into the realm of artificial intelligence, where an array of deep learning techniques has proven effective in automating crop leaf disease identification and classification. The current paper shows mature detection methodologies for apple, tomato, rice, mango, coconut, and durian leaf diseases with examples while demonstrating research on leaf disease detection in tropical plants. Through this exploration, valuable insights into the benefits and applications of detection techniques based on deep learning methods are provided for leaf disease detection. Highlighting the advantages of deep learning methods are provided for automated feature extraction and disease detection, the paper describes the salient features and challenges of the application of leaf disease detection in the tropics. In this paper, an introductory overview of a leaf disease detection model is offered and delve into the factors influencing detection accuracy and speed while proposing ways to mitigate the inherent trade-offs between these indicators. Furthermore, the challenges, such as multi-scale detection and leaf overlapping, that may occur in plants in the tropics, have been examined, enriching our understanding of deep learning-driven leaf disease detection in tropical agriculture.